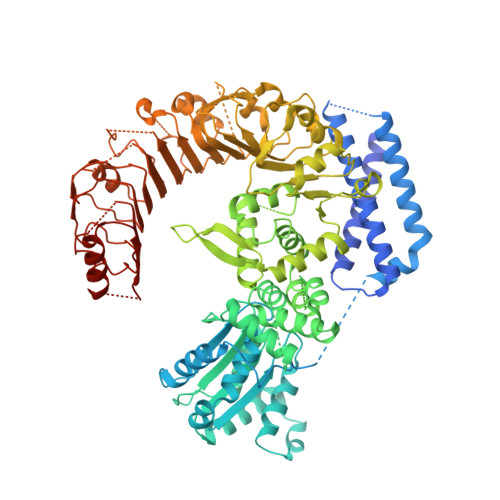
A disease resistance protein triggers oligomerization of its NLR helper into a hexameric resistosome to mediate innate immunity.
Madhuprakash, J., Toghani, A., Contreras, M.P., Posbeyikian, A., Richardson, J., Kourelis, J., Bozkurt, T.O., Webster, M.W., Kamoun, S.(2024) Sci Adv 10: eadr2594-eadr2594
- PubMed: 39504373
- DOI: https://doi.org/10.1126/sciadv.adr2594
- Primary Citation of Related Structures:
9FP6 - PubMed Abstract:
NRCs are essential helper NLR (nucleotide-binding domain and leucine-rich repeat) proteins that execute immune responses triggered by sensor NLRs. The resting state of NbNRC2 was recently shown to be a homodimer, but the sensor-activated state remains unclear. Using cryo-EM, we determined the structure of sensor-activated NbNRC2, which forms a hexameric inflammasome-like resistosome. Mutagenesis of the oligomerization interface abolished immune signaling, confirming the functional significance of the NbNRC2 resistosome. Comparative structural analyses between the resting state homodimer and sensor-activated homohexamer revealed substantial rearrangements, providing insights into NLR activation mechanisms. Furthermore, structural comparisons between NbNRC2 hexamer and previously reported CC-NLR pentameric assemblies revealed features allowing an additional protomer integration. Using the NbNRC2 hexamer structure, we assessed the recently released AlphaFold 3 for predicting activated CC-NLR oligomers, revealing high-confidence modeling of NbNRC2 and other CC-NLR amino-terminal α1 helices, a region proven difficult to resolve structurally. Overall, our work sheds light on NLR activation mechanisms and expands understanding of NLR structural diversity.
- The Sainsbury Laboratory, University of East Anglia, Norwich Research Park, Norwich, UK.
Organizational Affiliation: