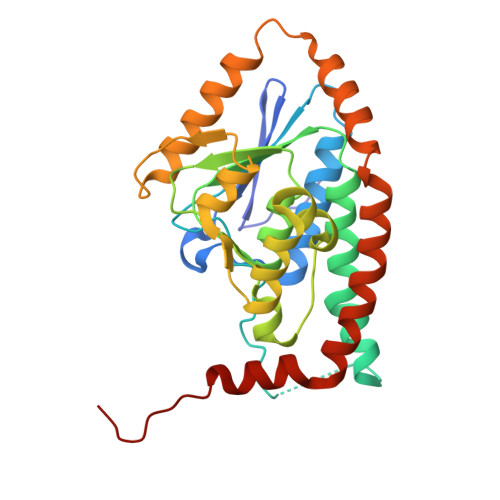
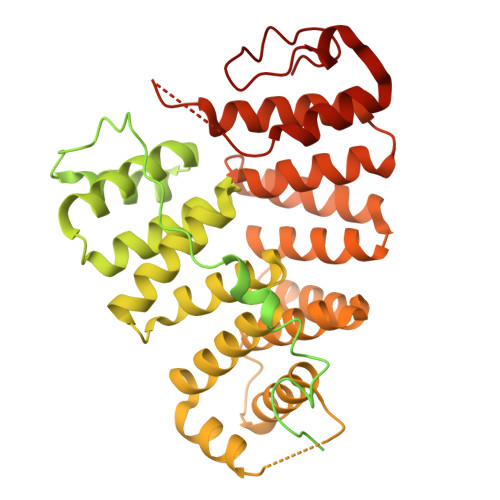
A cryo-electron microscopy structure of yeast Pex5 in complex with a cargo uncovers a novel binding interface.
Peer, L., Dym, O., Elad, N., Tirosh, A., Jacobovitch, J., Sivan, E., Angel, M., Albeck, S., Schuldiner, M., Peleg, Y., Zalckvar, E.(2025) J Cell Sci 138
- PubMed: 40376748
- DOI: https://doi.org/10.1242/jcs.263890
- Primary Citation of Related Structures:
9FGZ, 9FH0 - PubMed Abstract:
Proper protein targeting to organelles is crucial for maintaining eukaryotic cellular function and homeostasis. This necessity has driven the evolution of specific targeting signals on proteins and the targeting factors that recognize them. A prominent example is peroxisomal matrix proteins, most of which depend on the targeting factor Pex5 to localize and function correctly. Although most Pex5 cargoes contain a peroxisomal targeting signal type 1 (PTS1), they are not all targeted similarly. Some undergo priority targeting, facilitated either by stronger binding to specific subsets of PTS1 signals or by additional interaction interfaces. These observations highlight the extensive complexity of Pex5-mediated targeting. In this study, we reveal that the Saccharomyces cerevisiae (yeast) matrix protein Eci1 can reach peroxisomes and bind Pex5 in the absence of PTS1. By solving the structure of the yeast Pex5-Eci1 complex using cryo-electron microscopy, we identified additional binding interfaces. Our findings provide new insights into the versatile interactions between Pex5 and its cargo, Eci1. More broadly, this work highlights the intricate, dynamic nature of the interactions between cargo factors and their cargoes to meet the complex environment within eukaryotic cells.
- Department of Molecular Genetics, The Weizmann Institute of Science, Rehovot 7610001, Israel.
Organizational Affiliation: