CryoEM structure of human full-length alpha1beta3gamma2 GABA(A) receptor in complex with GARLH4, the TMD of Neuroligin2, GABA and Megabody38, in a desensitised state (StateD1)
Kasaragod, V.B., Aricescu, A.R.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gamma-aminobutyric acid receptor subunit alpha-1 | 413 | Homo sapiens | Mutation(s): 0 Gene Names: GABRA1 | 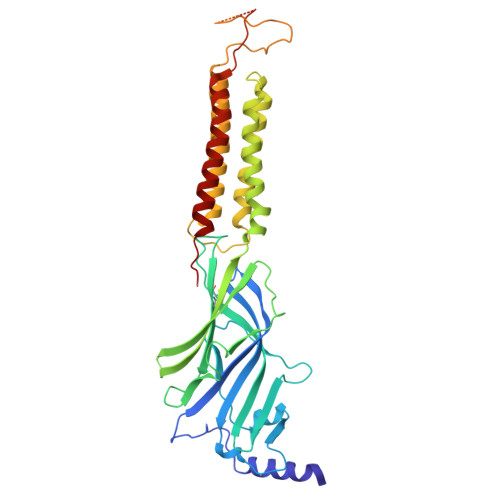 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P14867 (Homo sapiens) Explore P14867 Go to UniProtKB: P14867 | |||||
PHAROS: P14867 GTEx: ENSG00000022355 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P14867 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: P14867-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gamma-aminobutyric acid receptor subunit beta-3 | 441 | Homo sapiens | Mutation(s): 0 Gene Names: GABRB3 | 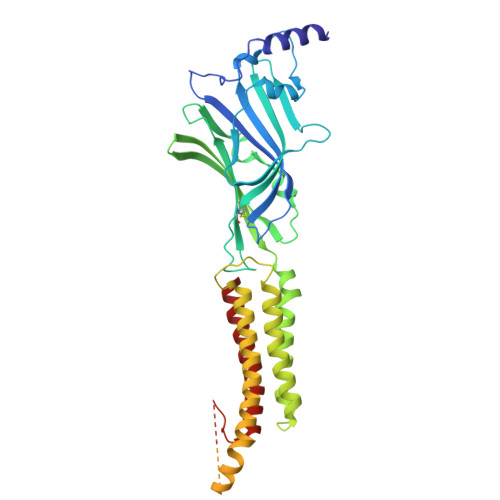 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P28472 (Homo sapiens) Explore P28472 Go to UniProtKB: P28472 | |||||
PHAROS: P28472 GTEx: ENSG00000166206 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P28472 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P28472-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Isoform 2 of Gamma-aminobutyric acid receptor subunit gamma-2 | 405 | Homo sapiens | Mutation(s): 0 Gene Names: GABRG2 | 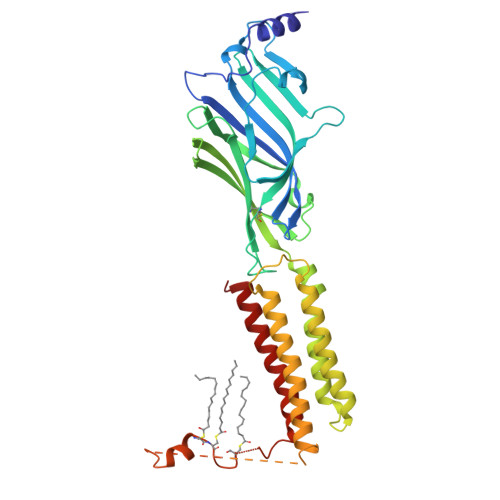 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P18507 (Homo sapiens) Explore P18507 Go to UniProtKB: P18507 | |||||
PHAROS: P18507 GTEx: ENSG00000113327 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P18507 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Neuroligin-2 | F [auth H] | 33 | Homo sapiens | Mutation(s): 0 Gene Names: NLGN2, KIAA1366 | 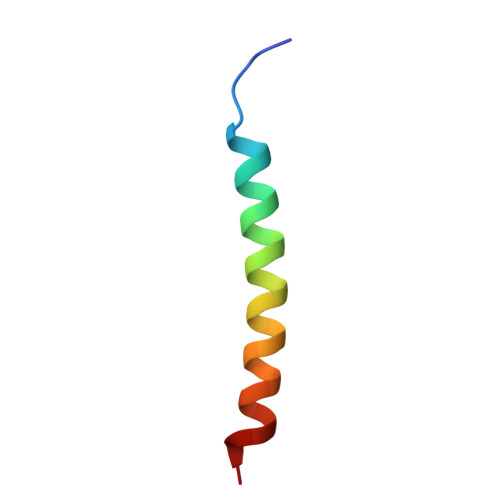 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8NFZ4 (Homo sapiens) Explore Q8NFZ4 Go to UniProtKB: Q8NFZ4 | |||||
PHAROS: Q8NFZ4 GTEx: ENSG00000169992 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8NFZ4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| LHFPL tetraspan subfamily member 4 protein | G [auth L] | 193 | Homo sapiens | Mutation(s): 0 Gene Names: LHFPL4, GARLH4 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q7Z7J7 (Homo sapiens) Explore Q7Z7J7 Go to UniProtKB: Q7Z7J7 | |||||
PHAROS: Q7Z7J7 GTEx: ENSG00000156959 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7Z7J7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Megabody38 | H [auth G], I [auth P] | 539 | Lama glama | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for B5Z8H1 (Helicobacter pylori (strain G27)) Explore B5Z8H1 Go to UniProtKB: B5Z8H1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B5Z8H1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | J [auth F] | 10 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G40702WU GlyCosmos: G40702WU GlyGen: G40702WU | |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | K [auth I], P [auth O] | 3 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G15407YE GlyCosmos: G15407YE GlyGen: G15407YE | |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | L [auth J], O [auth N] | 6 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G09724ZC GlyCosmos: G09724ZC GlyGen: G09724ZC | |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | M [auth K] | 2 |  | N/A | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | N [auth M] | 6 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G56014GC GlyCosmos: G56014GC GlyGen: G56014GC | |||||
| Ligands 11 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PGW Query on PGW | BB [auth E], IB [auth L], KB [auth L], R [auth A], SA [auth D] | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl
(9Z)-octadec-9-enoate C40 H77 O10 P PAZGBAOHGQRCBP-HGWHEPCSSA-N |  | ||
| PIO Query on PIO | Q [auth A], QA [auth D] | [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate C25 H49 O19 P3 XLNCEHRXXWQMPK-MJUMVPIBSA-N |  | ||
| PX2 Query on PX2 | IA [auth C], V [auth A], WA [auth D], YA [auth E], Z [auth B] | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE C27 H52 O8 P OKLASJZQBDJAPH-RUZDIDTESA-M |  | ||
| CLR (Subject of Investigation/LOI) Query on CLR | OA [auth C] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| OLC Query on OLC | KA [auth C] | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate C21 H40 O4 RZRNAYUHWVFMIP-GDCKJWNLSA-N |  | ||
| PLM Query on PLM | DB [auth E], HA [auth C], LA [auth C], MA [auth C], NA [auth C] | PALMITIC ACID C16 H32 O2 IPCSVZSSVZVIGE-UHFFFAOYSA-N |  | ||
| R16 Query on R16 | AA [auth B], EA [auth B], EB [auth E], ZA [auth E] | HEXADECANE C16 H34 DCAYPVUWAIABOU-UHFFFAOYSA-N |  | ||
| D10 Query on D10 | AB [auth E] BA [auth B] FB [auth E] GA [auth B] JA [auth C] | DECANE C10 H22 DIOQZVSQGTUSAI-UHFFFAOYSA-N |  | ||
| ABU (Subject of Investigation/LOI) Query on ABU | HB [auth E], W [auth A] | GAMMA-AMINO-BUTANOIC ACID C4 H9 N O2 BTCSSZJGUNDROE-UHFFFAOYSA-N |  | ||
| HEX Query on HEX | CA [auth B] CB [auth E] DA [auth B] FA [auth B] GB [auth E] | HEXANE C6 H14 VLKZOEOYAKHREP-UHFFFAOYSA-N |  | ||
| CL Query on CL | LB [auth L], PA [auth C], XA [auth D], Y [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| P1L Query on P1L | C | L-PEPTIDE LINKING | C19 H37 N O3 S |  | CYS |
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 19.2 |
| RECONSTRUCTION | RELION | 3.1 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MC_UP_1201/15 |
| European Molecular Biology Organization (EMBO) | European Union | ALTF137-2019 |
| H2020 Marie Curie Actions of the European Commission | European Union | GABAARComp-897707 |
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MR/L009609/1 |
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MC_EX_MR/T046279/1 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 1R01-GM135550 |
| National Science Foundation (NSF, United States) | United States | NeuroNex 2014862 |