Cryo-EM structure of the icosahedral lumazine synthase from Vicia faba.
Chee, M., Trapani, S., Hoh, F., Lai Kee Him, J., Yvon, M., Blanc, S., Bron, P.To be published.
Experimental Data Snapshot
Starting Model: other
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 6,7-dimethyl-8-ribityllumazine synthase | 157 | Vicia faba | Mutation(s): 0 EC: 2.5.1.78 | 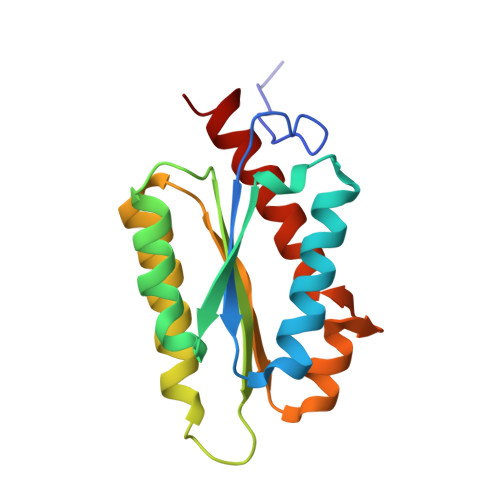 | |
UniProt | |||||
Find proteins for A0A9D4WQC0 (Pisum sativum) Explore A0A9D4WQC0 Go to UniProtKB: A0A9D4WQC0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A9D4WQC0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 4.0.0 |
| MODEL REFINEMENT | Coot | 0.9.8.92 |
| MODEL REFINEMENT | PHENIX | 1.20.1 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Montpellier University of Excellence (MUSE) | France | BLANC-MUSE2020-Multivir |