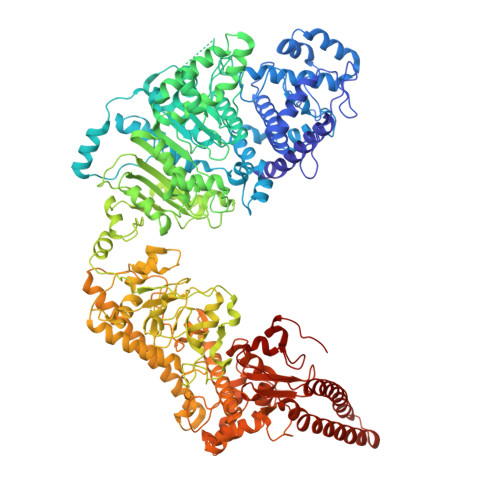
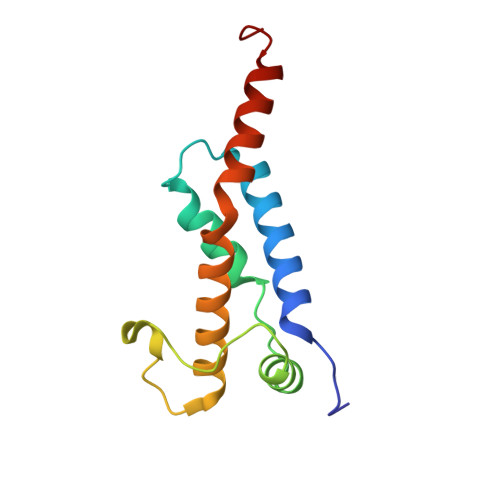
Molecular basis of foreign DNA recognition by BREX anti-phage immunity system.
Drobiazko, A., Adams, M.C., Skutel, M., Potekhina, K., Kotovskaya, O., Trofimova, A., Matlashov, M., Yatselenko, D., Maxwell, K.L., Blower, T.R., Severinov, K., Ghilarov, D., Isaev, A.(2025) Nat Commun 16: 1825-1825
- PubMed: 39979294
- DOI: https://doi.org/10.1038/s41467-025-57006-2
- Primary Citation of Related Structures:
9EWZ, 9EX7, 9EXH - PubMed Abstract:
Anti-phage systems of the BREX (BacteRiophage EXclusion) superfamily rely on site-specific epigenetic DNA methylation to discriminate between the host and invading DNA. We demonstrate that in Type I BREX systems, defense and methylation require BREX site DNA binding by the BrxX (PglX) methyltransferase employing S-adenosyl methionine as a cofactor. We determined 2.2-Å cryoEM structure of Escherichia coli BrxX bound to target dsDNA revealing molecular details of BREX DNA recognition. Structure-guided engineering of BrxX expands its DNA specificity and dramatically enhances phage defense. We show that BrxX alone does not methylate DNA, and BREX activity requires an assembly of a supramolecular BrxBCXZ immune complex. Finally, we present a cryoEM structure of BrxX bound to a phage-encoded inhibitor Ocr that sequesters BrxX in an inactive dimeric form. We propose that BrxX-mediated foreign DNA sensing is a necessary first step in activation of BREX defense.
- Skolkovo Institute of Science and Technology, Moscow, Russia.
Organizational Affiliation: