Novel citramalate lyase involved in glutamate fermentation of enterobacteria
Eggers, J., Schaefer, L., Cassens, E.A., Schmid, L., Simon, S.A., Probst, A.J., Koenig, S., Demmer, U., Ermler, U., Berg, I.A.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Citramalate lyase, subunit A (cmlA) | 456 | Klebsiella planticola | Mutation(s): 0 | 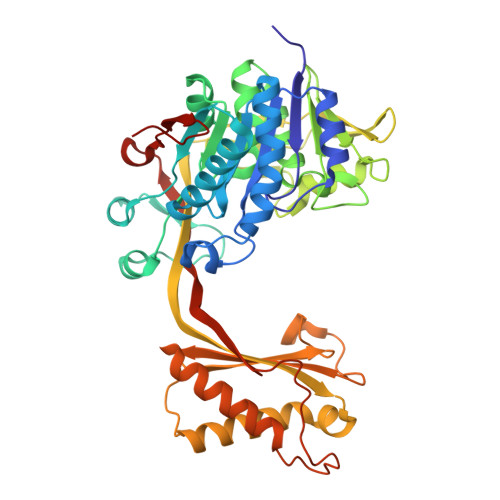 | |
UniProt | |||||
Find proteins for A0A0P8H7C5 (Citrobacter freundii) Explore A0A0P8H7C5 Go to UniProtKB: A0A0P8H7C5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0P8H7C5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Citramalate lyase, subunit B (cmlB) | 105 | Klebsiella planticola | Mutation(s): 0 | 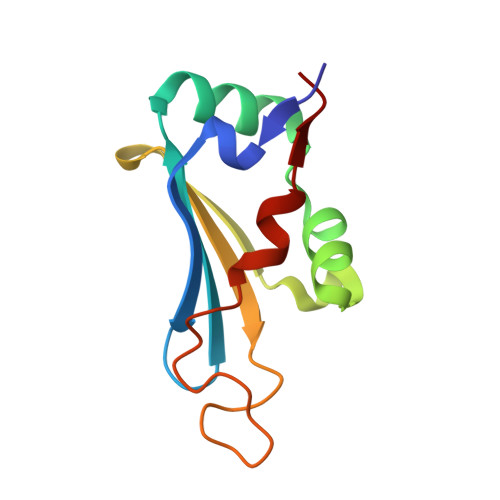 | |
UniProt | |||||
Find proteins for A0A564JVI2 (Klebsiella spallanzanii) Explore A0A564JVI2 Go to UniProtKB: A0A564JVI2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A564JVI2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP (Subject of Investigation/LOI) Query on ADP | E [auth A], L [auth C] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| MES Query on MES | I [auth A], J [auth A], P [auth C], Q [auth D] | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID C6 H13 N O4 S SXGZJKUKBWWHRA-UHFFFAOYSA-N |  | ||
| PYR (Subject of Investigation/LOI) Query on PYR | H [auth A], O [auth C] | PYRUVIC ACID C3 H4 O3 LCTONWCANYUPML-UHFFFAOYSA-N |  | ||
| CL Query on CL | K [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG (Subject of Investigation/LOI) Query on MG | F [auth A], G [auth A], M [auth C], N [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 107.21 | α = 90 |
| b = 108.34 | β = 90 |
| c = 98.55 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XSCALE | data scaling |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |