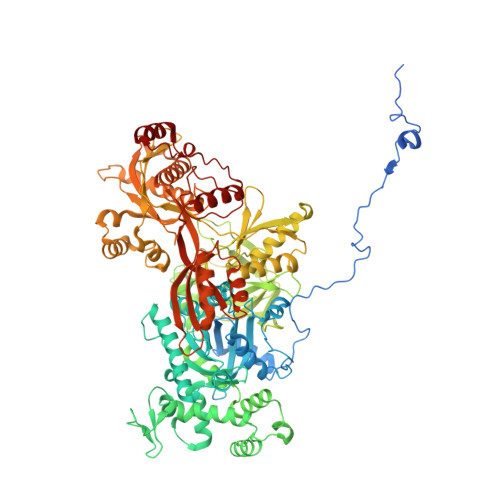
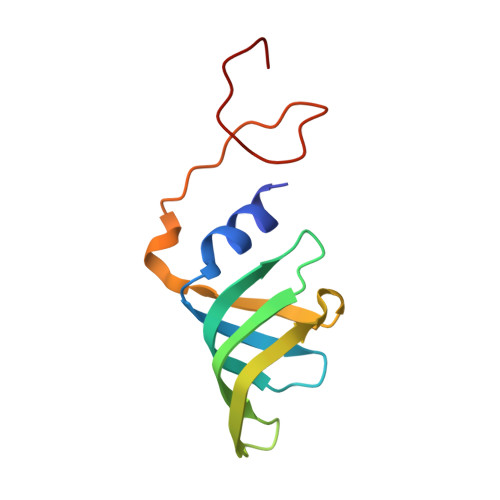
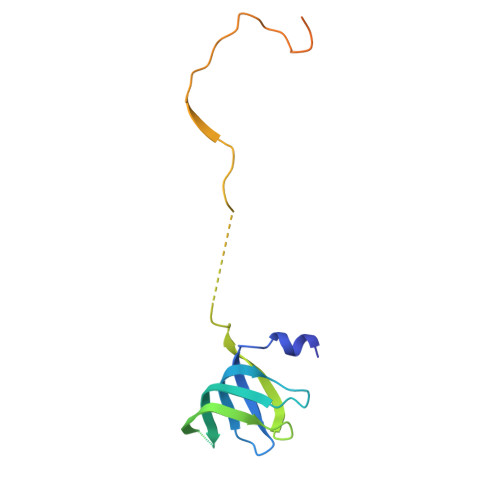
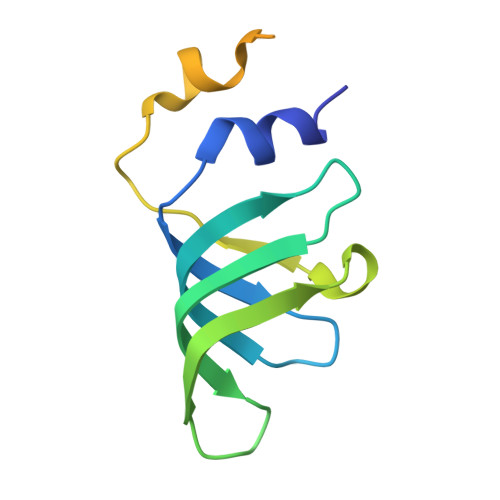
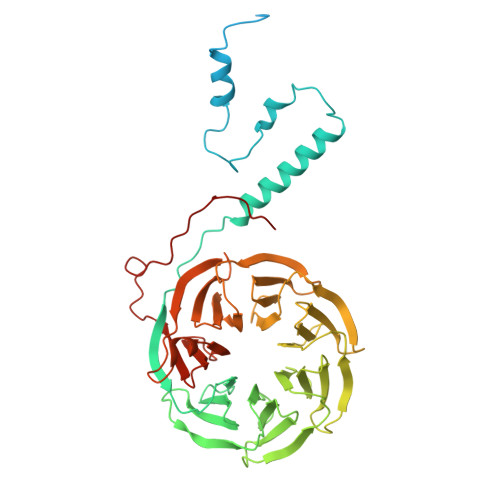
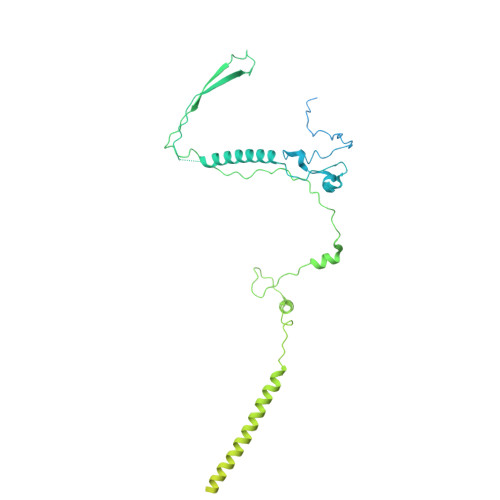
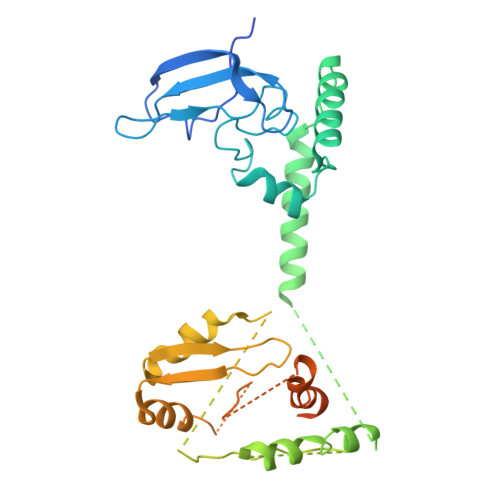
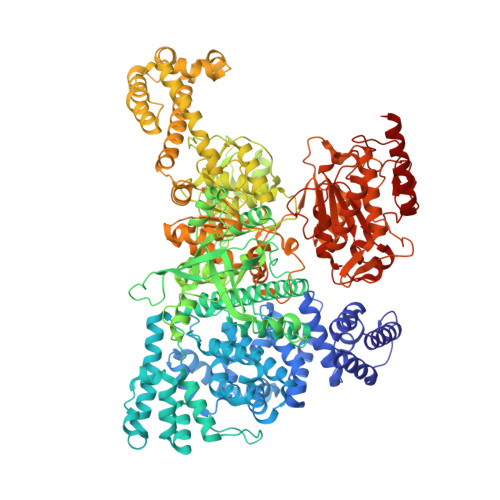
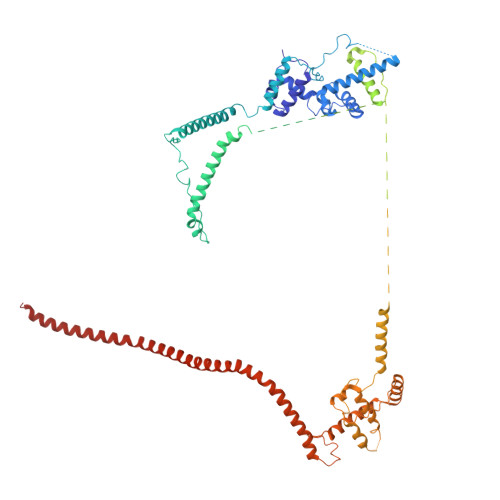
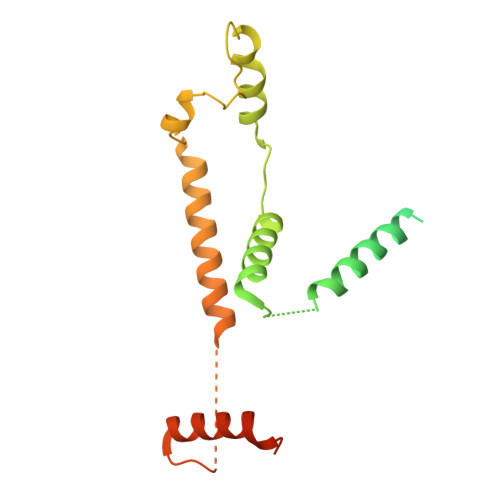
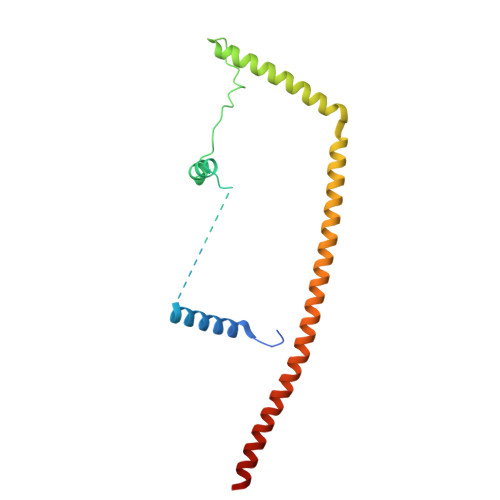
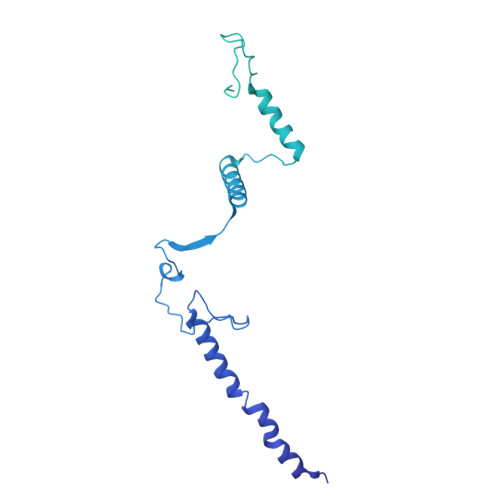
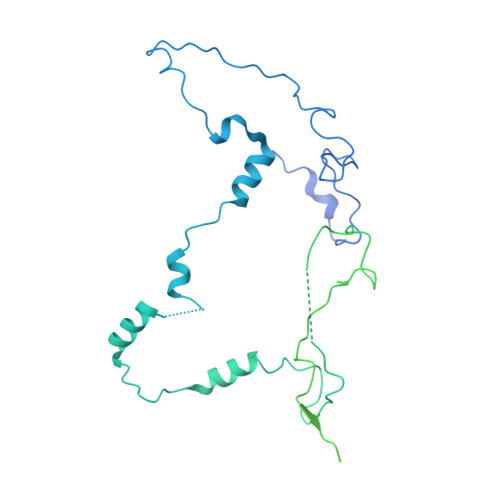
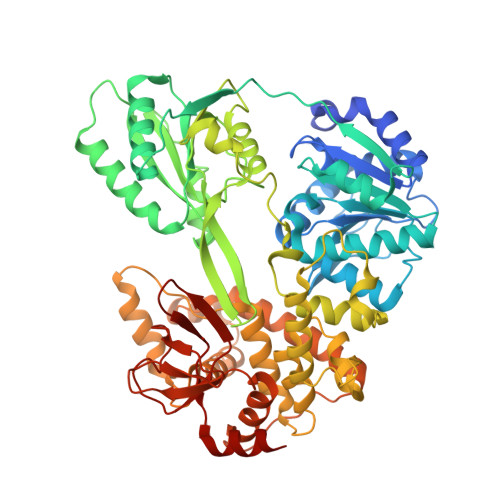
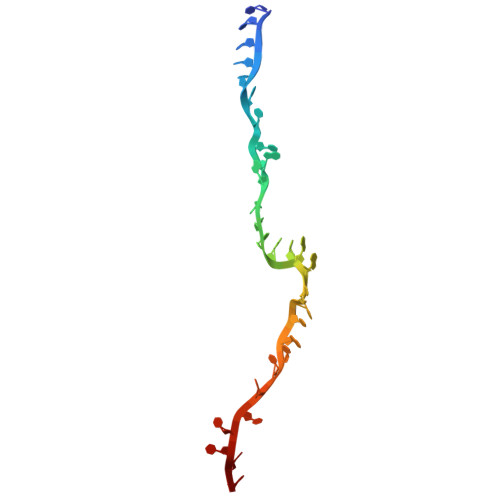
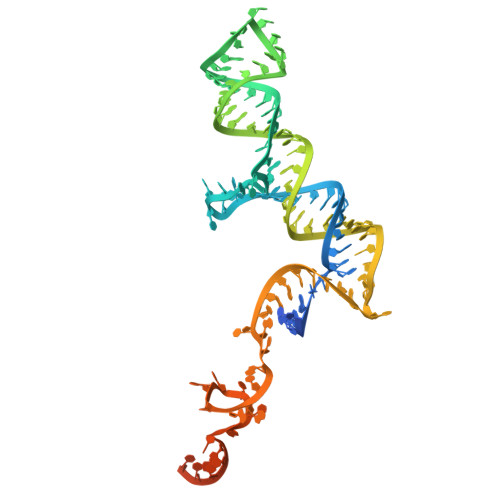
Structures of aberrant spliceosome intermediates on their way to disassembly.
Soni, K., Horvath, A., Dybkov, O., Schwan, M., Trakansuebkul, S., Flemming, D., Wild, K., Urlaub, H., Fischer, T., Sinning, I.(2025) Nat Struct Mol Biol 32: 914-925
- PubMed: 39833470
- DOI: https://doi.org/10.1038/s41594-024-01480-7
- Primary Citation of Related Structures:
9ESH, 9ESI - PubMed Abstract:
Intron removal during pre-mRNA splicing is of extraordinary complexity and its disruption causes a vast number of genetic diseases in humans. While key steps of the canonical spliceosome cycle have been revealed by combined structure-function analyses, structural information on an aberrant spliceosome committed to premature disassembly is not available. Here, we report two cryo-electron microscopy structures of post-B act spliceosome intermediates from Schizosaccharomyces pombe primed for disassembly. We identify the DEAH-box helicase-G-patch protein pair (Gih35-Gpl1, homologous to human DHX35-GPATCH1) and show how it maintains catalytic dormancy. In both structures, Gpl1 recognizes a remodeled active site introduced by an overstabilization of the U5 loop I interaction with the 5' exon leading to a single-nucleotide insertion at the 5' splice site. Remodeling is communicated to the spliceosome surface and the Ntr1 complex that mediates disassembly is recruited. Our data pave the way for a targeted analysis of splicing quality control.
- Heidelberg University Biochemistry Center (BZH), Heidelberg, Germany. komal.soni@uni-bayreuth.de.
Organizational Affiliation: