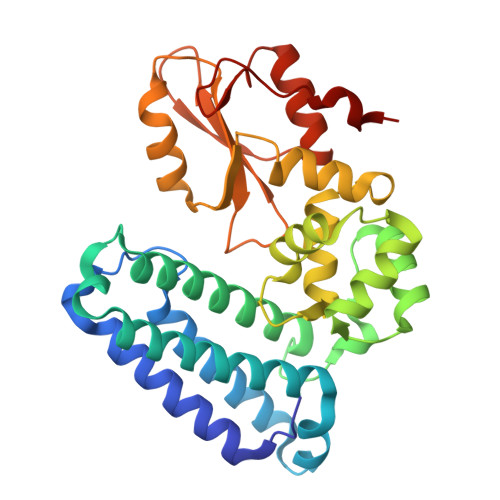
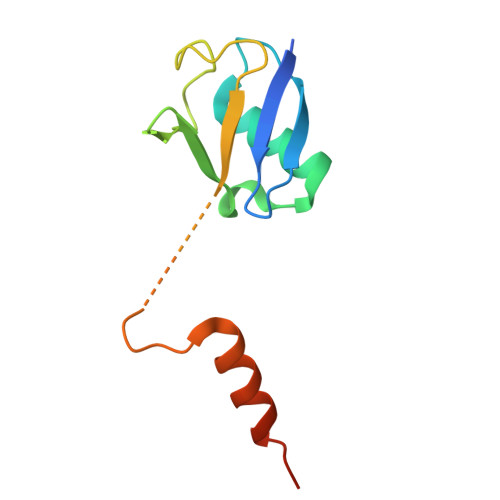
Locking CBL TKBD in its native conformation presents a novel therapeutic opportunity in mutant CBL-dependent leukemia.
Ahmed, S.F., Anand, J., Zhang, W., Buetow, L., Rishi, L., Mitchell, L., Bohlen, J., Lilla, S., Sibbet, G.J., Nixon, C., Patel, A., Majorek, K.A., Zanivan, S., Bustamante, J.C., Sidhu, S.S., Blyth, K., Huang, D.T.(2025) Mol Ther 33: 3624-3643
- PubMed: 40329529
- DOI: https://doi.org/10.1016/j.ymthe.2025.04.042
- Primary Citation of Related Structures:
9ERZ - PubMed Abstract:
Casitas B-lineage lymphoma (CBL) is an E3 ubiquitin ligase critical for negatively regulating receptor protein tyrosine kinases (RTKs). Deleterious CBL mutants lose E3 activity, but act as adaptors that gain function to cause myeloproliferative neoplasms. Currently, there is no targeted treatment available for patients with CBL mutant-dependent disorders. By combining phage-display technology and structure-based optimization, we discovered CBLock, a nanomolar affinity peptide inhibitor, that binds the substrate-binding site of CBL's tyrosine kinase binding domain (TKBD). CBLock disrupts the interaction between CBL mutants and RTKs, thereby impairing RTK-mediated priming of adaptor function of CBL mutants and downstream signaling. Notably, CBLock binds TKBD without inducing conformational changes, thereby preserving its ligand-free native conformation. In contrast, when CBL binds RTK substrates, TKBD undergoes a conformational change. Maintaining the native CBL TKBD conformation was crucial for CBLock to inhibit proliferation, induce cell cycle arrest, and promote apoptosis in leukemia cells harboring CBL mutations. In a mouse xenograft model of acute myeloid leukemia (AML), CBLock reduced tumor burden and improved survival rate. Moreover, CBLock inhibited the proliferation of cells derived from patients with CBL mutations. Therefore, inhibiting CBL TKBD in its native state presents a promising therapeutic opportunity in targeting mutant CBL-dependent leukemia.
- Cancer Research UK Scotland Institute, Garscube Estate, Switchback Road, Glasgow, G61 1BD, United Kingdom.
Organizational Affiliation: