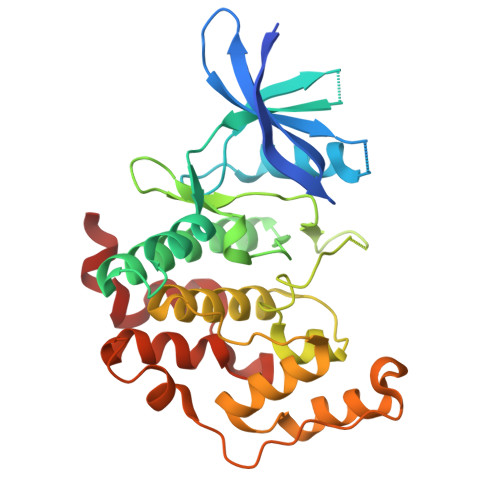
Crystal structure of the human CDKL5 kinase domain with compound CAF-382
Capener, J.L., Kim, J.K., Richardson, W., Silvaroli, J.A., Martinez, G.V., Thorson, V.C., Li, J., Hoyt, K.R., Satoskar, A.A., Mallipattu, S.K., Davidson, A.J., Zepeda-Orozco, D., Bajwa, A., Bullock, A.N., Axtman, A.D., Pabla, N.S.To be published.