Pseudomonas aeruginosa Cryptic Prophage Endolysin Is a Highly Active Muramidase.
Thoren Edvardsen, P.K., Englund, A.N., Kjendseth Rohr, A., Mesnage, S., Vaaje-Kolstad, G.(2025) Biochemistry 64: 3446-3458
- PubMed: 40631624
- DOI: https://doi.org/10.1021/acs.biochem.5c00142
- Primary Citation of Related Structures:
9EOI - PubMed Abstract:
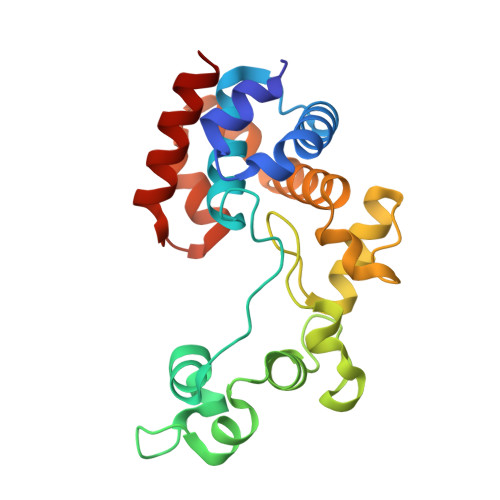
Endolysins are phage-encoded enzymes that cleave the peptidoglycan of host bacteria. These enzymes have gained considerable attention due to their ability to cause cell lysis, making them candidates as antibacterial agents. Most Pseudomonas aeruginosa genomes, including the common laboratory strains PAO1 and UCBPP-PA14, contain a cryptic prophage encoding a glycoside hydrolase family 19 endolysin (named Pa GH19Lys in the present study). Family 19 glycoside hydrolases are known to target peptidoglycan and chitin-type substrates. Pa GH19Lys was not active toward chitin but exhibited activity toward chloroform-treated Gram-negative bacteria, displaying ∼10,000-fold higher activity than hen egg white lysozyme. Analysis of products derived from Pa GH19Lys activity toward purified P. aeruginosa peptidoglycan showed that the enzyme catalyzed hydrolysis of the β-1,4 linkage between N- acetylmuramic acid and N- acetyl-d-glucosamine, classifying the enzyme as a muramidase. Finally, the crystal structure of Pa GH19Lys was determined and solved to 1.8 Å resolution. The structure of the enzyme showed a globular α-helical fold possessing a deep but relatively open catalytic cleft.
- Faculty of Chemistry, Biotechnology, and Food Science, Norwegian University of Life Sciences, Ås 1432, Norway.
Organizational Affiliation: