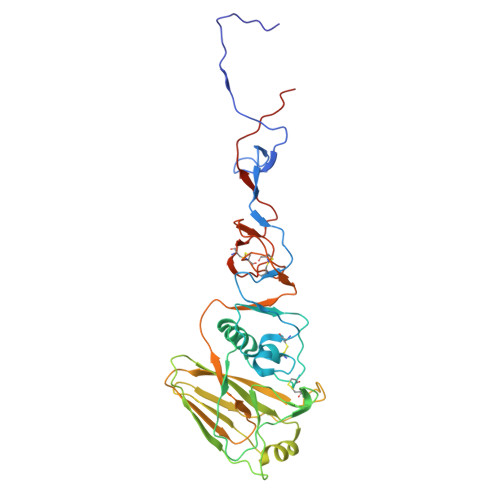
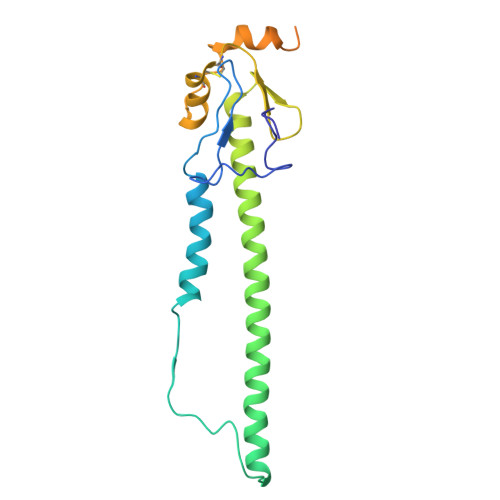
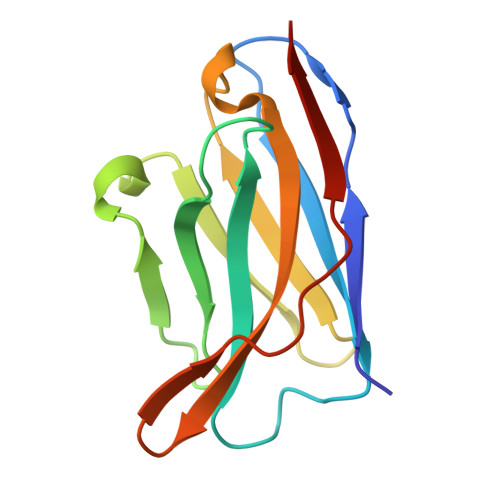
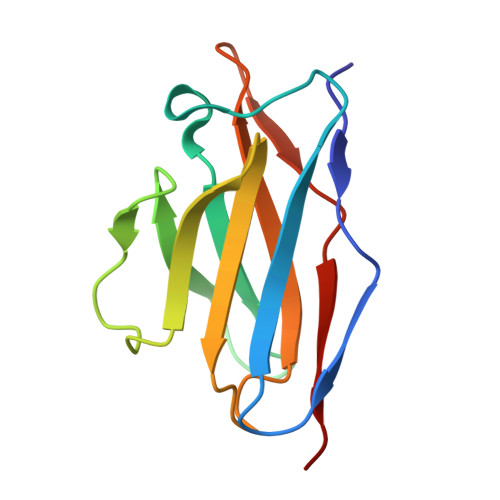
The N terminus of H3-influenza hemagglutinin as a site-of-vulnerability to neutralizing antibody.
Rawi, R., Morano, N.C., Cheung, C.S., Du, H., Gorman, J., Prabhakaran, M., Becker, J.E., Bylund, T., Charaf, S., Chen, X., Lee, M., Harris, D.R., Olia, A.S., Ou, L., Wang, L., Wang, S., Zhang, B., Kanekiyo, M., McDermott, A.B., Zhou, T., Shapiro, L., Kwong, P.D.(2025) Structure 33: 1820
- PubMed: 40816277
- DOI: https://doi.org/10.1016/j.str.2025.07.015
- Primary Citation of Related Structures:
9E69, 9EI8, 9EI9 - PubMed Abstract:
The N terminus of the H3 subtype of influenza virus hemagglutinin is ∼10 residues longer than the N termini of most other hemagglutinins. As conserved, exposed, and linear regions may be good vaccine targets, we investigated the vaccine utility of the extended H3-N terminus. First, we identified antibody 5E10, for which structure and binding analyses revealed recognition of the H3-N terminus. Second, we immunized mice with immunogens incorporating the H3-N terminus, boosted with hemagglutinin trimer, and isolated antibodies from immunogen-elicited B cells that bound both H3-N terminus and hemagglutinin trimer. However, hemagglutinin-complex structures of two such antibodies, 3864-6 and 3864-10, that neutralized H3-influenza strains, revealed only peripheral recognition of the hemagglutinin N terminus. Collectively, these results reveal the N terminus of H3 hemagglutinin to be a suboptimal vaccine target and suggest that-in addition to being conserved, flexible, and accessible-other factors influence the elicitation of potent broadly neutralizing responses.
- Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA. Electronic address: reda.rawi@nih.gov.
Organizational Affiliation: