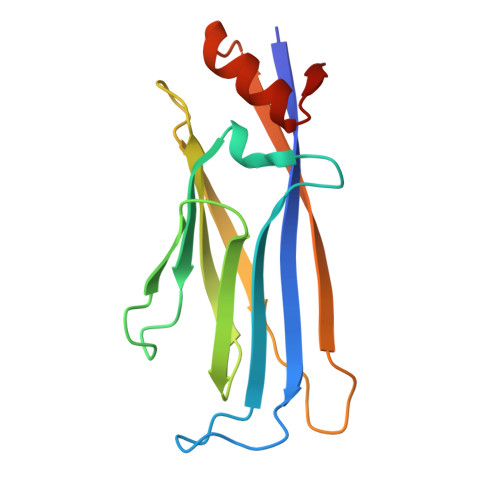
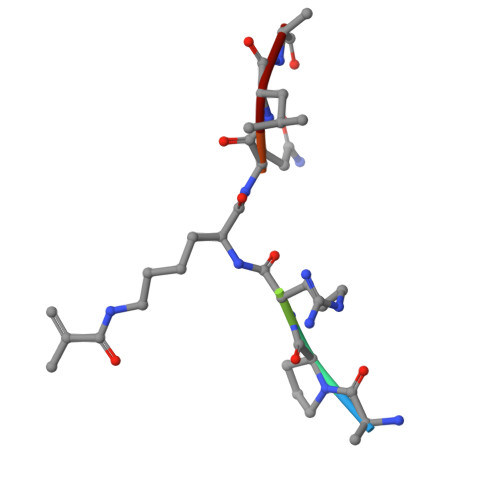
The YEATS domain is a selective reader of histone methacrylation.
Becht, D.C., Song, J., Selvam, K., Yin, K., Bai, W., Zhao, Y., Wu, R., Zheng, Y.G., Kutateladze, T.G.(2025) Structure 33: 1233-1239.e5
- PubMed: 40339582
- DOI: https://doi.org/10.1016/j.str.2025.04.010
- Primary Citation of Related Structures:
9EHI - PubMed Abstract:
Metabolically regulated lysine acylation modifications in proteins play a major role in epigenetic processes and cellular homeostasis. A new type of histone acylation, lysine methacrylation, has recently been identified but remains poorly characterized. Here, we show that lysine methacrylation can be generated through metabolism of sodium methacrylate and enzymatically removed in cells, and that the YEATS domain but not bromodomain recognizes this modification. Structural and biochemical analyses reveal the π-π-π-stacking mechanism for binding of the YEATS domain of ENL to methacrylated histone H3K18 (H3K18mc). Using mass spectrometry proteomics, we demonstrate that methacrylate induces global methacrylation of a set of proteins that differs from the set of methacrylated proteins associated with valine metabolism. These findings suggest that high levels of methacrylate may potentially perturb cellular functions of these proteins by altering protein methacrylation profiles.
- Department of Pharmacology, University of Colorado School of Medicine, Aurora, CO 80045, USA.
Organizational Affiliation: