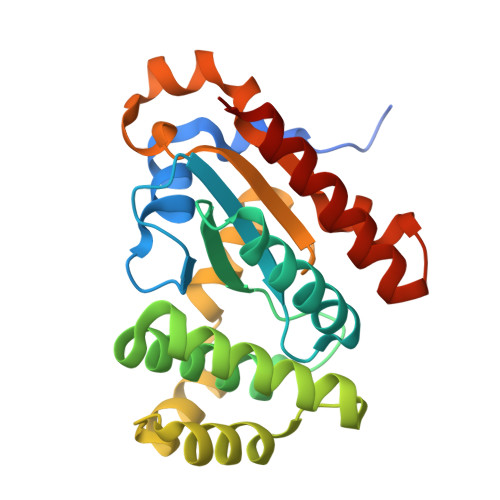
Unveiling the versatility of the thioredoxin framework: Insights from the structural examination of Francisella tularensis DsbA1.
Penning, S., Hong, Y., Cunliffe, T., Hor, L., Totsika, M., Paxman, J.J., Heras, B.(2024) Comput Struct Biotechnol J 23: 4324-4336
- PubMed: 39697679
- DOI: https://doi.org/10.1016/j.csbj.2024.11.034
- Primary Citation of Related Structures:
9EDL - PubMed Abstract:
In bacteria the formation of disulphide bonds is facilitated by a family of enzymes known as the disulphide bond forming (Dsb) proteins, which, despite low sequence homology, belong to the thioredoxin (TRX) superfamily. Among these enzymes is the disulphide bond-forming protein A (DsbA); a periplasmic thiol oxidase responsible for catalysing the oxidative folding of numerous cell envelope and secreted proteins. Pathogenic bacteria often contain diverse Dsb proteins with distinct functionalities commonly associated with pathogenesis. Here we investigate FtDsbA1, a DsbA homologue from the Gram-negative bacterium Francisella tularensis . Our study shows that FtDsbA1 shares a conserved TRX-like fold bridged by an alpha helical bundle showcased by all DsbA-like proteins. However, FtDsbA1 displays a highly unique variation on this structure, containing an extended and flexible N-terminus and secondary structural elements inserted within the core of the TRX fold itself, which together twist the overall DsbA-like architecture. Additionally, FtDsbA1 exhibits variations to the well conserved active site with an unusual dipeptide in the catalytic CXXC redox centre (CGKC), and a trans configuration for the conserved cis -proline loop, known for governing DsbA-substrate interactions. FtDsbA1's redox properties are comparable to other DsbA enzymes, however, consistent with its atypical structure, functional analysis reveals FtDsbA1 has a high degree of substrate specificity suggesting a specialised role within F. tularensis' oxidative folding pathway. Overall, this work underscores the remarkable malleability of the TRX catalytic core, a ubiquitous and ancestral protein fold. This not only contributes to broadening the structural and functional diversity seen within proteins utilising this core fold but will also enhance the accuracy of AI-driven protein structural prediction tools.
- Department of Biochemistry and Chemistry, La Trobe Institute for Molecular Science, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, Australia.
Organizational Affiliation: