Structural Basis for Target Discrimination and Activation by Cas13d
Chou, C.W., Finkelstein, I.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| EsCas13d | 954 | [Eubacterium] siraeum DSM 15702 | Mutation(s): 0 Gene Names: EUBSIR_02687 | 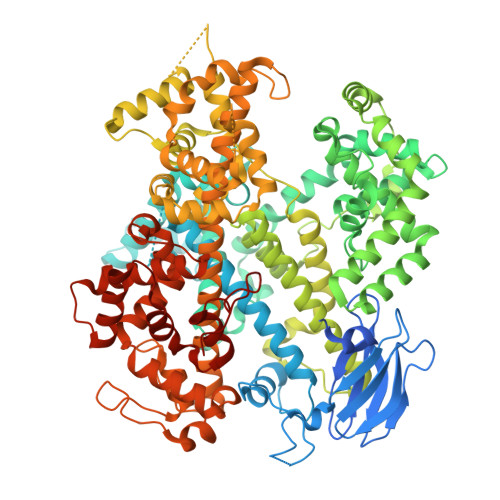 | |
UniProt | |||||
Find proteins for B0MS50 ([Eubacterium] siraeum DSM 15702) Explore B0MS50 Go to UniProtKB: B0MS50 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B0MS50 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| crRNA | 52 | [Eubacterium] siraeum DSM 15702 | 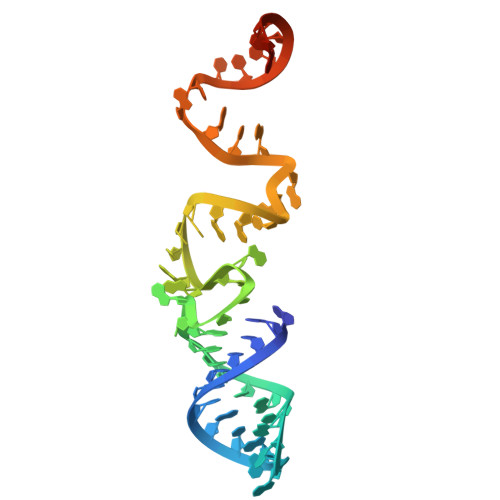 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MG Query on MG | C [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21.2_5419 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Welch Foundation | United States | F-1808 |