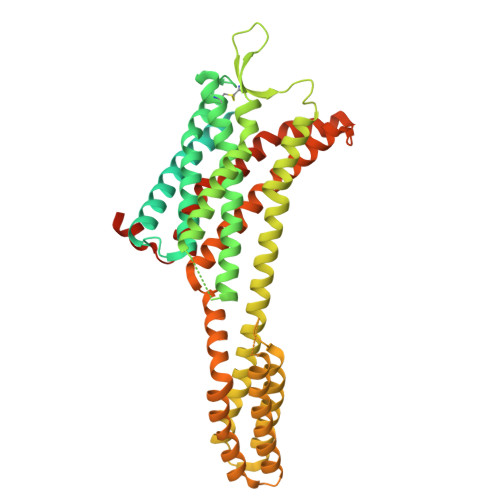
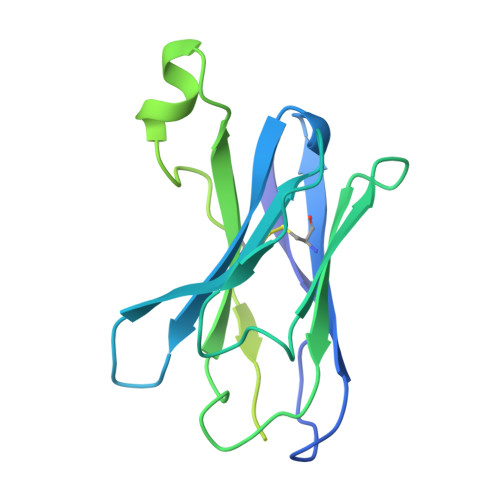
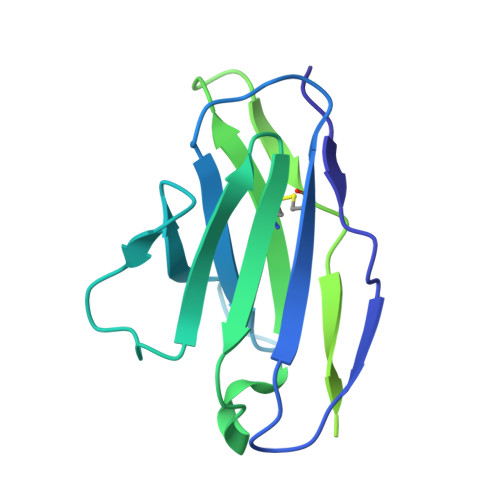
Epitope-directed selection of GPCR nanobody ligands with evolvable function.
Skiba, M.A., Canavan, C., Nemeth, G.R., Liu, J., Kanso, A., Kruse, A.C.(2025) Proc Natl Acad Sci U S A 122: e2423931122-e2423931122
- PubMed: 40067891
- DOI: https://doi.org/10.1073/pnas.2423931122
- Primary Citation of Related Structures:
9EAH, 9EAI, 9EAJ - PubMed Abstract:
Antibodies have the potential to target G protein-coupled receptors (GPCRs) with high receptor, cellular, and tissue selectivity; however, few antibody ligands for GPCRs exist. Here, we describe a generalizable selection method to enrich for GPCR ligands from a synthetic camelid antibody fragment (nanobody) library. Our strategy yielded multiple nanobody ligands for the angiotensin II type I receptor (AT1R), a prototypical GPCR and important drug target. We found that nanobodies readily act as allosteric modulators, encoding selectivity for both the receptor and chemical features of GPCR ligands. We then used structure-guided design to convert two nanobodies from allosteric ligands to competitive AT1R inhibitors through simple mutations. This work demonstrates that nanobodies can encode multiple pharmacological behaviors and have great potential as evolvable scaffolds for the development of next-generation GPCR therapeutics.
- Department of Biological Chemistry and Molecular Pharmacology, Blavatnik Institute, Harvard Medical School, Boston, MA 02115.
Organizational Affiliation: