Cryo-EM structure of inactive and active prostaglandin D2 receptor
Davoudinasab, B., Han, G.W., Cherezov, V.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | 394 | Homo sapiens | Mutation(s): 8 Gene Names: GNAS, GNAS1, GSP EC: 3.6.5 Membrane Entity: Yes | 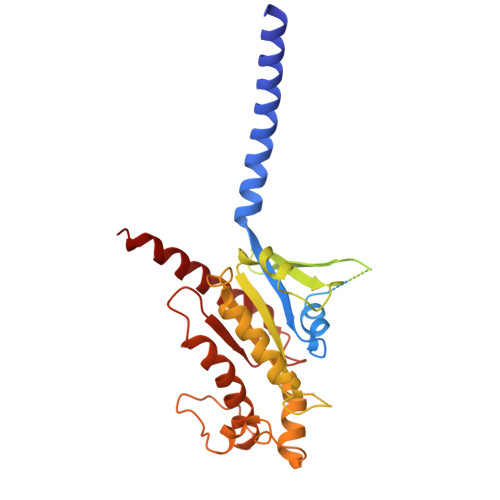 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P63092 (Homo sapiens) Explore P63092 Go to UniProtKB: P63092 | |||||
PHAROS: P63092 GTEx: ENSG00000087460 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63092 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | 345 | Homo sapiens | Mutation(s): 0 Gene Names: GNB1 | 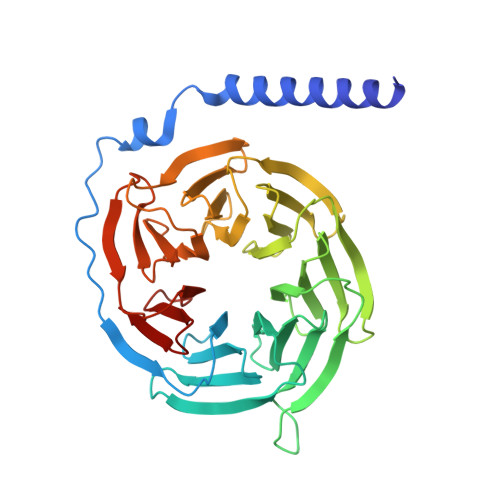 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62873 (Homo sapiens) Explore P62873 Go to UniProtKB: P62873 | |||||
PHAROS: P62873 GTEx: ENSG00000078369 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62873 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 | 71 | Homo sapiens | Mutation(s): 0 Gene Names: GNG2 Membrane Entity: Yes | 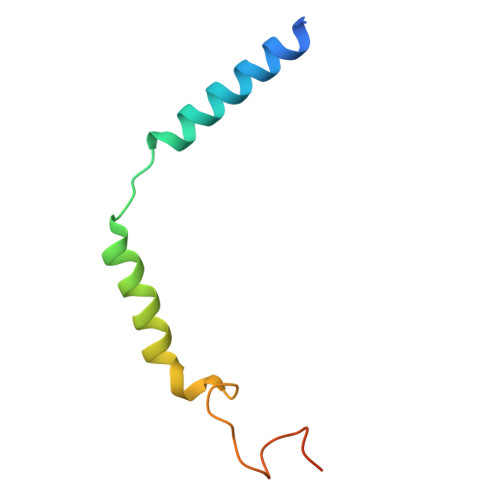 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P59768 (Homo sapiens) Explore P59768 Go to UniProtKB: P59768 | |||||
PHAROS: P59768 GTEx: ENSG00000186469 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P59768 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Prostaglandin D2 receptor | 385 | Homo sapiens | Mutation(s): 0 Gene Names: PTGDR Membrane Entity: Yes | 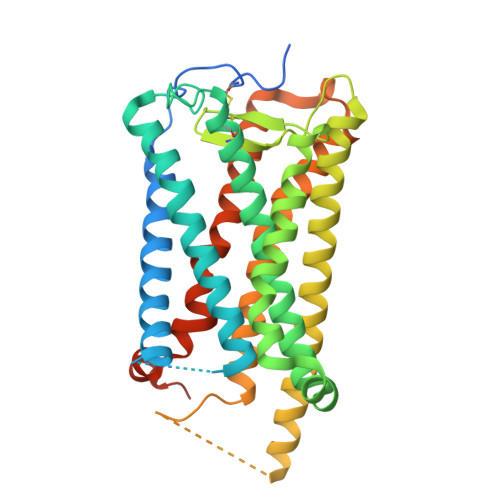 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q13258 (Homo sapiens) Explore Q13258 Go to UniProtKB: Q13258 | |||||
PHAROS: Q13258 GTEx: ENSG00000168229 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q13258 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: Q13258-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nanobody35|Lama glama | E [auth N] | 139 | Lama glama | Mutation(s): 0 | 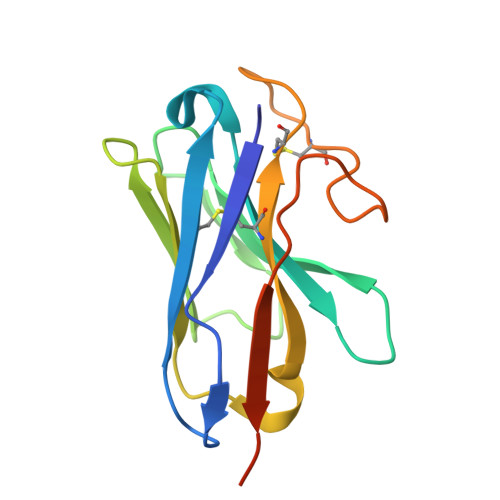 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| Y01 Query on Y01 | H [auth D] | CHOLESTEROL HEMISUCCINATE C31 H50 O4 WLNARFZDISHUGS-MIXBDBMTSA-N |  | ||
| PG2 (Subject of Investigation/LOI) Query on PG2 | F [auth D] | PROSTAGLANDIN D2 C20 H32 O5 BHMBVRSPMRCCGG-OUTUXVNYSA-N |  | ||
| NAG Query on NAG | G [auth D] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R35GM127086 |