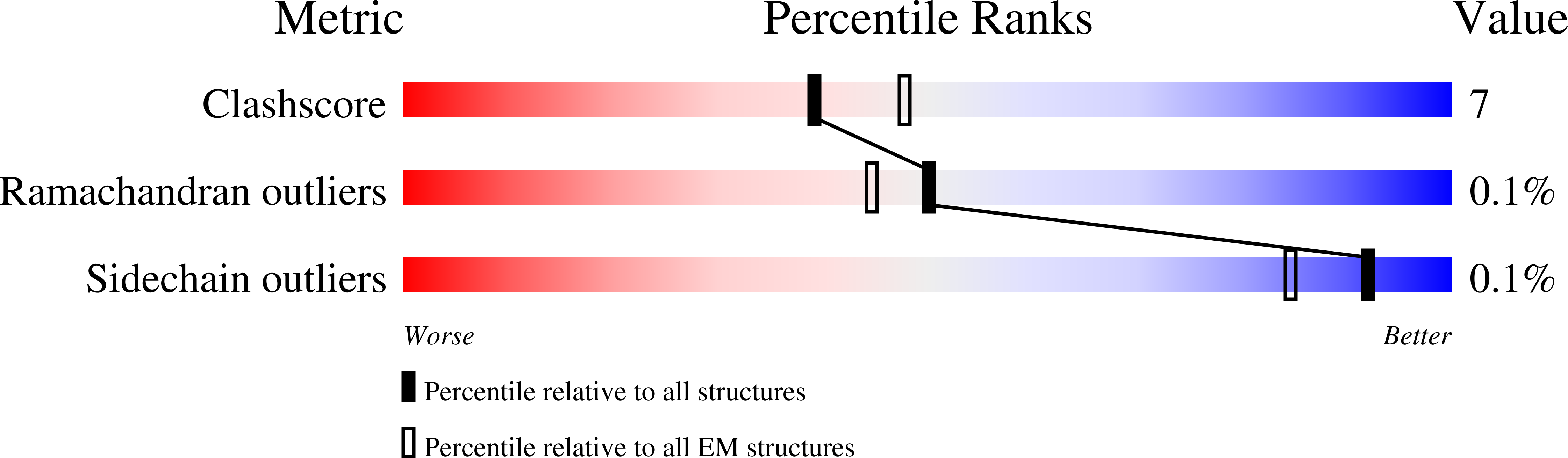Memantine inhibits calcium-permeable AMPA receptors.
Carrillo, E., Montano Romero, A., Gonzalez, C.U., Turcu, A.L., Vazquez, S., Twomey, E.C., Jayaraman, V.(2025) Nat Commun 16: 5576-5576
- PubMed: 40593487
- DOI: https://doi.org/10.1038/s41467-025-60543-5
- Primary Citation of Related Structures:
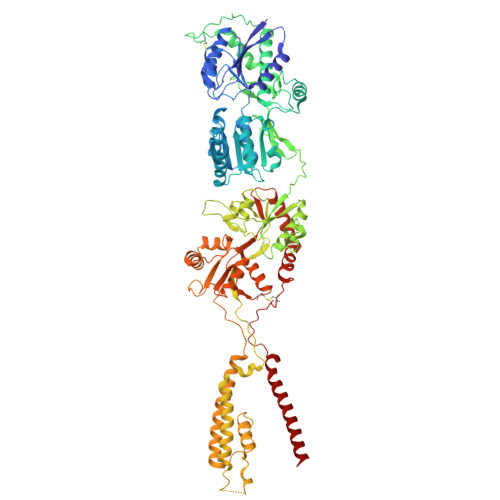
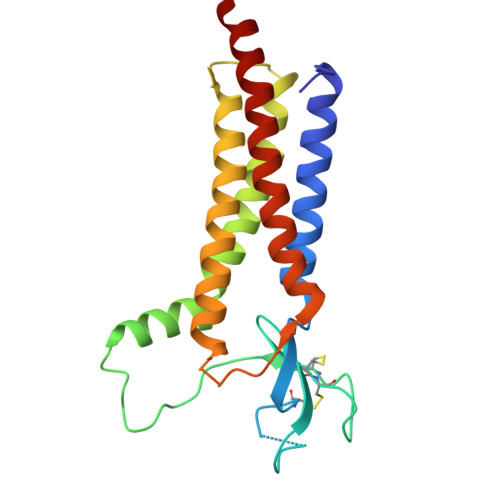
9E4Y, 9E4Z - PubMed Abstract:
Memantine is an US Food and Drug Administration (FDA) approved drug that is thought to selectively inhibit NMDA-subtype of ionotropic glutamate receptors (NMDARs). NMDARs enable calcium influx into neurons and are critical for normal brain function. However, increasing evidence shows that calcium influx in neurological diseases is augmented by calcium-permeable AMPA-subtype ionotropic glutamate receptors (AMPARs). Here, we demonstrate that these calcium-permeable AMPARs (CP-AMPARs) are inhibited by memantine. Electrophysiology unveils that memantine inhibition of CP-AMPARs is dependent on their calcium permeability and the presence of their neuronal auxiliary subunit transmembrane AMPAR regulatory proteins (TARPs). Through cryo-electron microscopy we elucidate that memantine blocks CP-AMPAR ion channels in a unique mechanism of action from NMDARs. Furthermore, we demonstrate that memantine inhibits a gain of function AMPAR mutation found in a patient with a neurodevelopmental disorder. Our findings unlock potential exploitation of this site to design more specific drugs targeting CP-AMPARs.
- Center for Membrane Biology, Department of Biochemistry and Molecular Biology, University of Texas Health Science Center at Houston, Houston, TX, USA. elisa.carrilloflores@uth.tmc.edu.
Organizational Affiliation: