Robust light-harvesting properties upon low-light acclimation in purple bacteria
Wang, D., Harris, D., Chuang, C., Schmidt, G.P., Fiebig, O.C., Schlau-Cohen, G.S.(2025) Chem 11: 102597
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2025) Chem 11: 102597
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Light-harvesting protein B-800/820 alpha chain | 48 | Rhodoblastus acidophilus | Mutation(s): 0 | 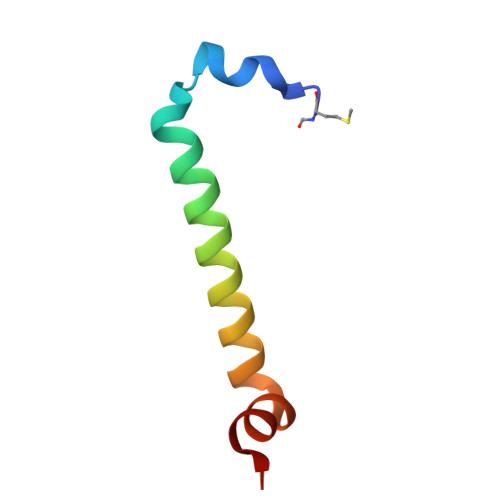 | |
UniProt | |||||
Find proteins for P35090 (Rhodoblastus acidophilus) Explore P35090 Go to UniProtKB: P35090 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P35090 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Light-harvesting protein B-800/820 beta-2 chain | 41 | Rhodoblastus acidophilus | Mutation(s): 0 | 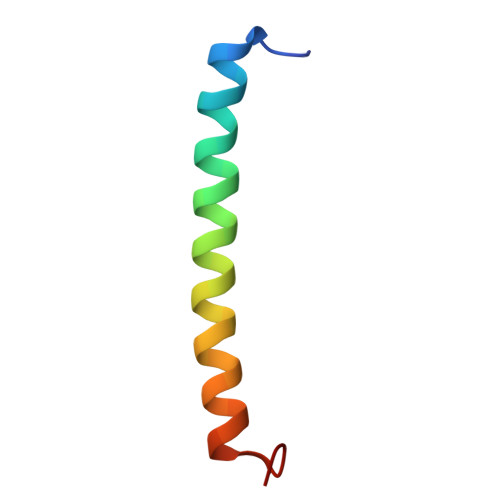 | |
UniProt | |||||
Find proteins for P35096 (Rhodoblastus acidophilus) Explore P35096 Go to UniProtKB: P35096 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P35096 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BCL (Subject of Investigation/LOI) Query on BCL | AB [auth L] CA [auth D] CB [auth M] DB [auth M] GA [auth E] | BACTERIOCHLOROPHYLL A C55 H74 Mg N4 O6 DSJXIQQMORJERS-AGGZHOMASA-M |  | ||
| IRM (Subject of Investigation/LOI) Query on IRM | BA [auth C] FA [auth E] FB [auth M] JB [auth O] NA [auth G] | 1,2-Dihydro-psi,psi-caroten-1-ol C40 H58 O CNYVJTJLUKKCGM-RGGGOQHISA-N |  | ||
| LMT Query on LMT | BB [auth L] DA [auth D] HB [auth N] JA [auth F] NB [auth P] | DODECYL-BETA-D-MALTOSIDE C24 H46 O11 NLEBIOOXCVAHBD-QKMCSOCLSA-N |  | ||
| LDA Query on LDA | AA [auth C] EA [auth E] EB [auth M] IB [auth O] MA [auth G] | LAURYL DIMETHYLAMINE-N-OXIDE C14 H31 N O SYELZBGXAIXKHU-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| FME Query on FME | A, C, E, G, I A, C, E, G, I, K, M, O, Q | L-PEPTIDE LINKING | C6 H11 N O3 S |  | MET |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Department of Energy (DOE, United States) | United States | DE-SC0018097 |
| National Science Foundation (NSF, United States) | United States | DMR-1419807 |