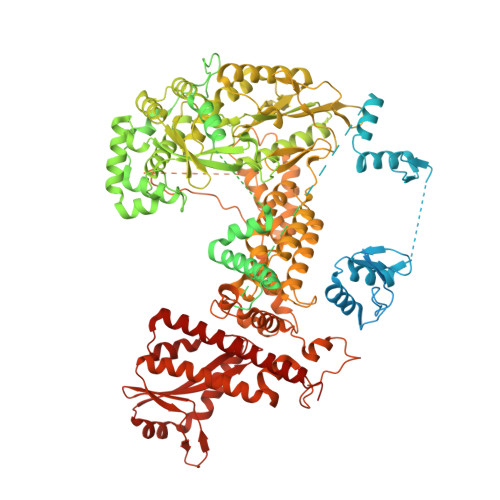
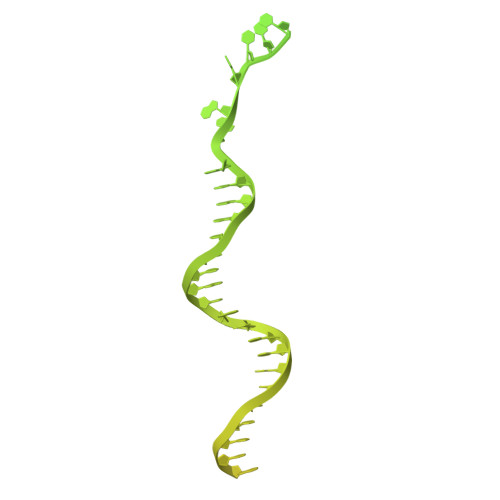
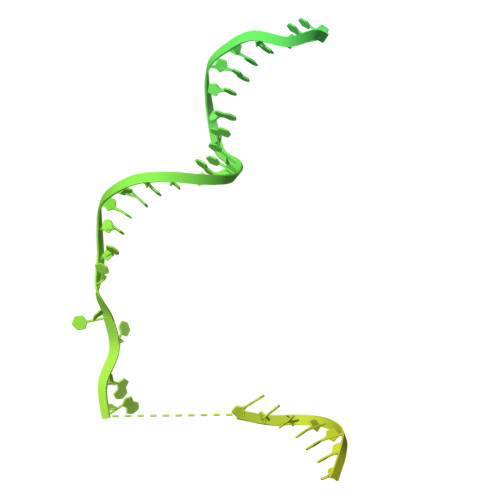
Structure and biochemistry-guided engineering of an all-RNA system for DNA insertion with R2 retrotransposons.
Edmonds, K.K., Wilkinson, M.E., Strebinger, D., Chen, H., Lash, B., Schaefer, C.C., Zhu, S., Liu, D., Zilberzwige-Tal, S., Ladha, A., Walsh, M.L., Frangieh, C.J., Vaz Reay, N.A., Macrae, R.K., Wang, X., Zhang, F.(2025) Nat Commun 16: 6079-6079
- PubMed: 40603868
- DOI: https://doi.org/10.1038/s41467-025-61321-z
- Primary Citation of Related Structures:
9DOU - PubMed Abstract:
R2 elements, a class of non-long terminal repeat (non-LTR) retrotransposons, have the potential to be harnessed for transgene insertion. However, efforts to achieve this are limited by our understanding of the retrotransposon mechanisms. Here, we structurally and biochemically characterize R2 from Taeniopygia guttata (R2Tg). We show that R2Tg cleaves both strands of its ribosomal DNA target and binds a pseudoknotted RNA element within the R2 3' UTR to initiate target-primed reverse transcription. Guided by these insights, we engineer and characterize an all-RNA system for transgene insertion. We substantially reduce the system's size and insertion scars by eliminating unnecessary R2 sequences on the donor. We further improve the integration efficiency by chemically modifying the 5' end of the donor RNA and optimizing delivery, creating a compact system that achieves over 80% integration efficiency in several human cell lines. This work expands the genome engineering toolbox and provides mechanistic insights that will facilitate future development of R2-mediated gene insertion tools.
- Howard Hughes Medical Institute, Cambridge, MA, USA.
Organizational Affiliation: