DnaB and DciA: mechanisms of helicase loading and translocation on ssDNA.
Gao, N., Mazzoletti, D., Peng, A., Olinares, P.D.B., Morrone, C., Garavaglia, A., Gouda, N., Tsoy, S., Mendoza, A., Chowdhury, A., Cerullo, A., Bhavsar, H., Rossi, F., Rizzi, M., Chait, B.T., Miggiano, R., Jeruzalmi, D.(2025) Nucleic Acids Res 53
- PubMed: 40598899
- DOI: https://doi.org/10.1093/nar/gkaf521
- Primary Citation of Related Structures:
9DLS - PubMed Abstract:
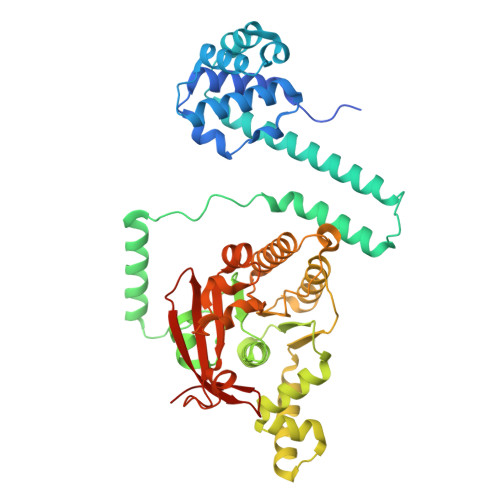
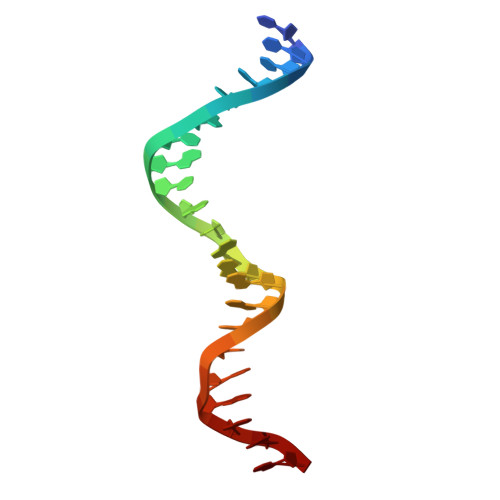
Replicative helicases are assembled on chromosomes by helicase loaders before the initiation of DNA replication. Here, we investigate the mechanisms employed by the bacterial Vibrio cholerae (Vc) DnaB replicative helicase and the DciA helicase loader. Structural analysis of the ATPγS form of the VcDnaB-ssDNA complex reveals a configuration distinct from that observed with GDP•AlF4. With ATPγS, the amino-terminal domain (NTD) tier, previously found as an open spiral in the GDP•AlF4 complex, adopts a closed planar arrangement. Furthermore, the DnaB subunit at the top of the carboxy-terminal domain (CTD) spiral is displaced by approximately 25 Å between the two forms. We suggest that remodeling the NTD layer between closed planar and open spiral configurations, along with the migration of two distinct CTDs to the top of the DnaB spiral, repeated three times, mediates hand-over-hand translocation. Biochemical analysis indicates that VcDciA utilizes its Lasso domain to interact with DnaB near its Docking-Helix Linker-Helix interface. Up to three copies of VcDciA bind to VcDnaB, suppressing its ATPase activity during loading onto physiological DNA substrates. Our data suggest that DciA loads DnaB onto DNA using the ring-opening mechanism.
- Department of Chemistry and Biochemistry, City College of New York, New York, NY 10031, United States.
Organizational Affiliation: