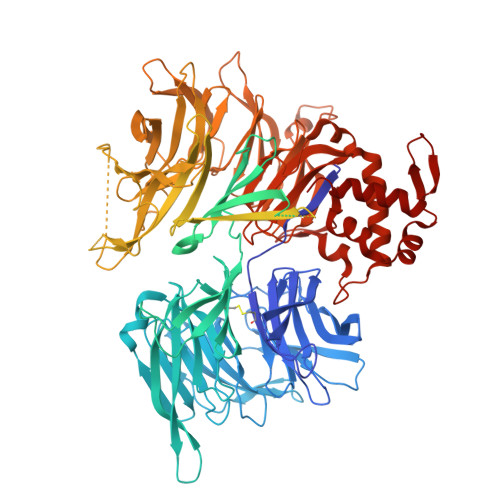
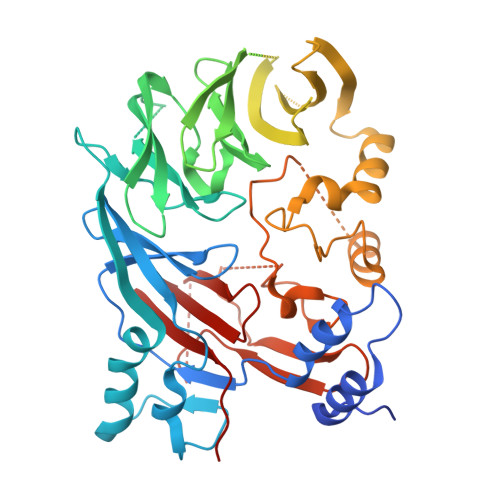
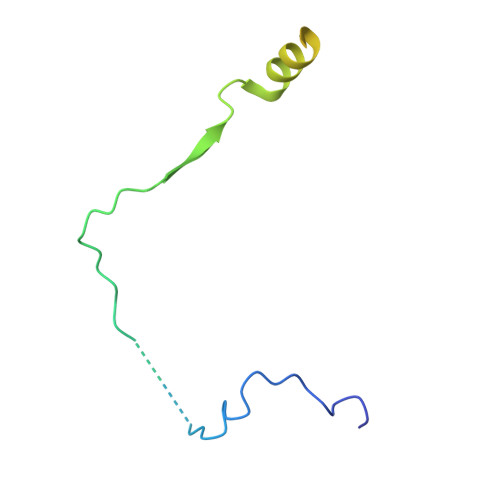
Impairment of DET1 causes neurological defects and lethality in mice and humans.
Karayel, O., Soung, A., Gurung, H., Schubert, A.F., Klaeger, S., Kschonsak, M., Al-Maraghi, A., Bhat, A.A., Alshabeeb Akil, A.S., Dugger, D.L., Webster, J.D., French, D.M., Anand, D., Soni, N., Fakhro, K.A., Rose, C.M., Harris, S.F., Ndoja, A., Newton, K., Dixit, V.M.(2025) Proc Natl Acad Sci U S A 122: e2422631122-e2422631122
- PubMed: 39937864
- DOI: https://doi.org/10.1073/pnas.2422631122
- Primary Citation of Related Structures:
9DHD - PubMed Abstract:
COP1 and DET1 are components of an E3 ubiquitin ligase that is conserved from plants to humans. Mammalian COP1 binds to DET1 and is a substrate adaptor for the CUL4A-DDB1-RBX1 RING E3 ligase. Transcription factor substrates, including c-Jun, ETV4, and ETV5, are targeted for proteasomal degradation to effect rapid transcriptional changes in response to cues such as growth factor deprivation. Here, we link a homozygous DET1 R26W mutation to lethal developmental abnormalities in humans. Experimental cryo-electron microscopy of the DET1 complex with DDB1 and DDA1, as well as co-immunoprecipitation experiments, revealed that DET1 R26W impairs binding to DDB1, thereby compromising E3 ligase function. Accordingly, human-induced pluripotent stem cells homozygous for DET1 R26W expressed ETV4 and ETV5 highly, and exhibited defective mitochondrial homeostasis and aberrant caspase-dependent cell death when differentiated into neurons. Neuronal cell death was increased further in the presence of Det1 -deficient microglia as compared to WT microglia, indicating that the deleterious effects of the DET1 p.R26W mutation may stem from the dysregulation of multiple cell types. Mice lacking Det1 died during embryogenesis, while Det1 deletion just in neural stem cells elicited hydrocephalus, cerebellar dysplasia, and neonatal lethality. Our findings highlight an important role for DET1 in the neurological development of mice and humans.
- Department of Physiological Chemistry, Genentech, South San Francisco, CA 94080.
Organizational Affiliation: