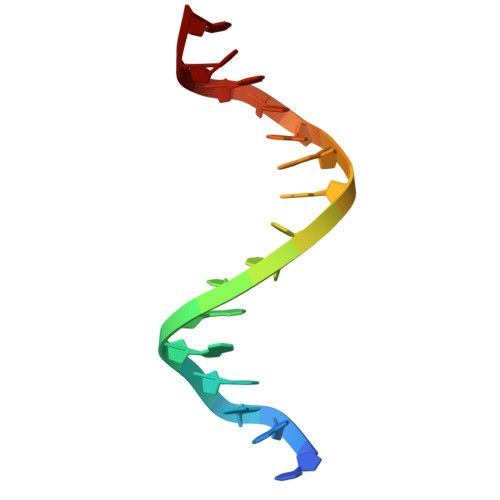
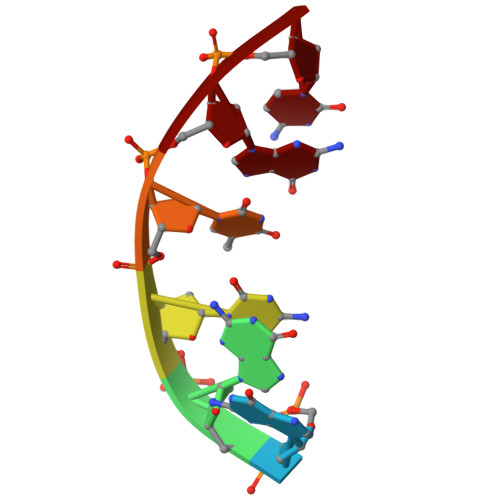
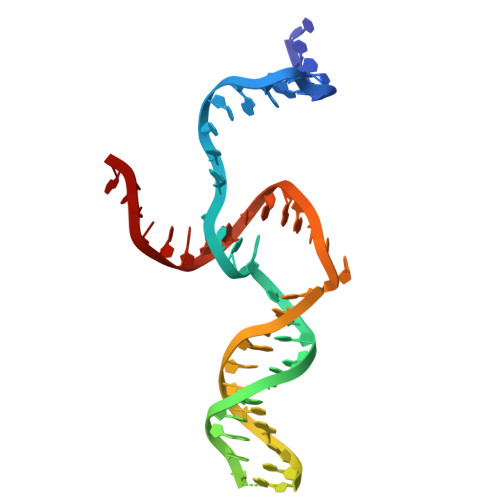
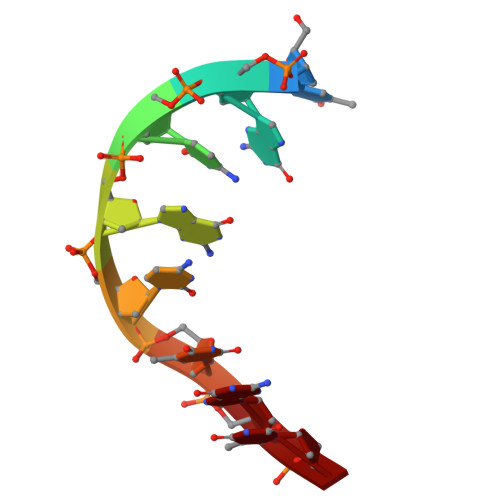
Crystallographic Study of DNA T-Junction via Crystal Engineering.
Li, X., Siraj, N., Sha, R., Mao, C.(2025) Angew Chem Int Ed Engl 64: e18174-e18174
- PubMed: 41165523
- DOI: https://doi.org/10.1002/anie.202518174
- Primary Citation of Related Structures:
9DFY, 9DFZ, 9DG0, 9DGL, 9DGM, 9DGN - PubMed Abstract:
Engineering DNA crystals is the primary motivation for structural DNA nanotechnology. Among many potential applications, such crystals promise as a platform to precisely (in terms of both location and orientation) organize biomolecules into 3D crystals for X-ray crystallographic studies of the guest biomolecules. The crystal formation depends on rationally designed DNA frameworks instead of unpredictable interactions between the guest molecule themselves; thus, avoiding the crystallization problem of biomolecules. This approach was proposed 40 years ago, however, has not been realized so far. Herein, we report an effort along this direction to study DNA T-junction, a common DNA structure used in DNA nanoconstruction. This study is an initial demonstration of the feasibility of the 40-year-old proposal. We expect that it would be quickly adapted to study many other molecules, particularly nucleic acids (e.g., aptamers, catalytic DNAs/RNAs, ribozymes, and ribonucleoproteins) that are, otherwise, difficult to be studied.
- Purdue University, Department of Chemistry, West Lafayette, IN, 47907, USA.
Organizational Affiliation: