Is a Malleable Active Site Loop the Key to High Substrate Promiscuity? Hybrid, Biocatalytic Route to Structurally Diverse Taxoid Side Chains with Remarkable Dual Stereocontrol.
Kudalkar, G.P., Leidner, F., Kumar, N., Hass, J.L., Madzelan, P., Powell, D.R., Day, V.W., Magueres, P.L., Ferrara, J.D., Daniels, L.M., Yamano, A., Ito, S., Niu, W., Grubmuller, H., Wilson, M.A., Berkowitz, D.B.(2025) Angew Chem Int Ed Engl 64: e202510889-e202510889
- PubMed: 40537411
- DOI: https://doi.org/10.1002/anie.202510889
- Primary Citation of Related Structures:
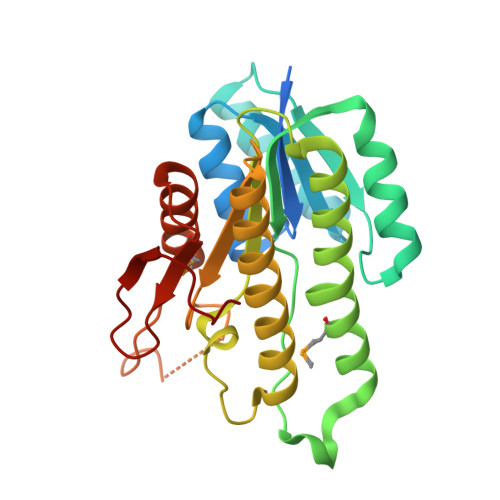
9D7J, 9NYO - PubMed Abstract:
These studies reveal the first structure of Clostridiumacetobutylicum alcohol dehydrogenase (CaADH), a protein exhibiting remarkable substrate promiscuity and stereochemical fidelity. The CaADH enzyme is utilized here for synthesizing 20 potential aryl isoserine side chains for the Taxotère family of tubulin-binding chemotherapeutics. The approach involves dynamic reductive kinetic resolution (DYRKR) upon the corresponding α-chloro-β-keto esters, showing high D-syn stereoselectivity, including those leading to the clinically relevant Milataxel (Ar = 2-furyl) and Simotaxel (Ar = 2-thienyl) side chains. Furthermore, various cross-coupling chemistries performed on the p-bromophenyl isoserine side chain significantly enhance the structural diversity of the taxoid side chain library obtained (16 additional taxoid side chains). The CaADH structure is notable: (i) the nicotinamide cofactor is bound in an anti-conformation, with the amide carbonyl occupying the ketone binding pocket, and (ii) a flexible loop near the active site, likely contributes to the remarkable substrate promiscuity observed in CaADH. We present our perspective on the dynamic nature of the CaADH active site through molecular dynamics simulation, proposing a halogen bonding model as a potential mechanism for the remarkable selectivity for an (S)-configured C-Cl bond, in addition to the D-facial selectivity, demonstrated across twenty diverse substrates by this remarkable short-chain dehydrogenase enzyme.
- University of Nebraska-Lincoln, Chemistry, Department of Chemistry, 68588-0304, Lincoln, UNITED STATES OF AMERICA.
Organizational Affiliation: