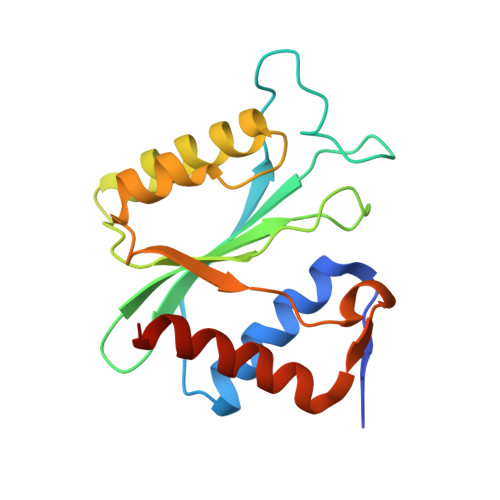
Asymmetric loading of TnsE regulates Tn7 targeting of DNA replication structures.
Krishnan, S.S., Shen, Y., O'Hagan, T.B., Matthews, L.A., Weerasinghe, N.W., Ghirlando, R., Thibodeaux, C.J., Guarne, A.(2025) Nucleic Acids Res 53
- PubMed: 40498074
- DOI: https://doi.org/10.1093/nar/gkaf472
- Primary Citation of Related Structures:
9D5L - PubMed Abstract:
Tn7 transposable elements are known for their sophisticated target-site selection mechanisms. For the prototypical Tn7 element, dedicated transposon-encoded proteins direct insertions to either a conserved site in the chromosome or replicating DNA structures in conjugal plasmids, ensuring the vertical and horizontal spread of the element. While the pathway targeting the attTn7 site in the bacterial chromosome has been extensively studied, the pathway targeting DNA replication structures remains poorly understood. We have used an integrative structural biology approach to elucidate how the Tn7-encoded protein TnsE recognizes replication sites. Using native mass spectrometry, we found that TnsE forms 1:1 and 2:1 (TnsE:DNA) complexes on 3'-recessed DNA, with gain-of-function TnsE variants favoring the formation of 2:1 complexes. Structural characterization confirms that two TnsE molecules bind to DNA with the C-terminal domain of the protein recognizing duplex DNA, leaving the N-terminal domain to impose DNA substrate specificity and recruit the core transposition machinery. Collectively, our work is consistent with a model where TnsE-mediated target-site selection relies on the formation of an asymmetric TnsE:DNA complex to recruit the Tn7 transposase to DNA replication structures.
- Department of Biochemistry, McGill University, Montreal, QC H3G 0B1, Canada.
Organizational Affiliation: