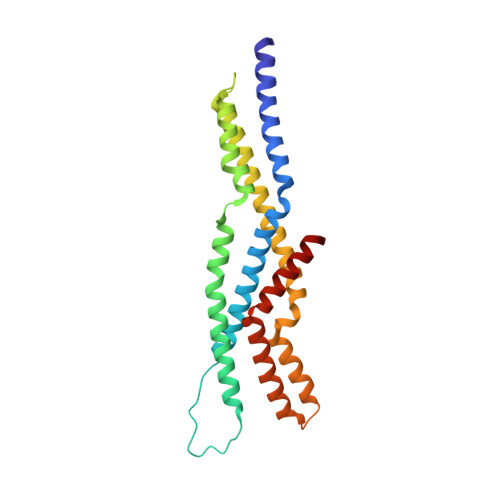
Fungal kinesin-8 motors dimerize by folding their proximal tail domain into a compact helical bundle.
Trofimova, D., Doubleday, C., Hunter, B., Serrano Arevalo, J.D., Davison, E., Wen, E., Munro, K., Allingham, J.S.(2025) Structure 33: 2122
- PubMed: 40907488
- DOI: https://doi.org/10.1016/j.str.2025.08.011
- Primary Citation of Related Structures:
9CRW - PubMed Abstract:
Kinesin-8 motors regulate kinetochore-microtubule dynamics and control spindle length and positioning. Certain isoforms achieve this by traversing microtubules, accumulating at plus-ends, and depolymerizing terminal αβ-tubulin subunits. While the kinesin-8 motor domain is well characterized, the tail domain regions are less understood. Using the Candida albicans Kip3 protein as a model for fungal kinesin-8, we present an X-ray crystal structure and hydrodynamic analysis of its motor-proximal tail segment, revealing its role in motor dimerization. This segment forms a compact, 92 Å-long four-helix bundle, rather than an elongated coiled-coil stalk seen in most kinesins. The bundle is stabilized primarily by interactions between helices one and three, with additional support from helices two and four. A flexible hinge bisects the bundle into two lobules, imparting mechanical pliability and asymmetric exterior surfaces. These unique features may facilitate interactions with regulatory elements or contribute to the functional versatility of kinesin-8 motors.
- Department of Biomedical and Molecular Sciences, Queen's University, Kingston, ON K7L 3N6, Canada.
Organizational Affiliation: