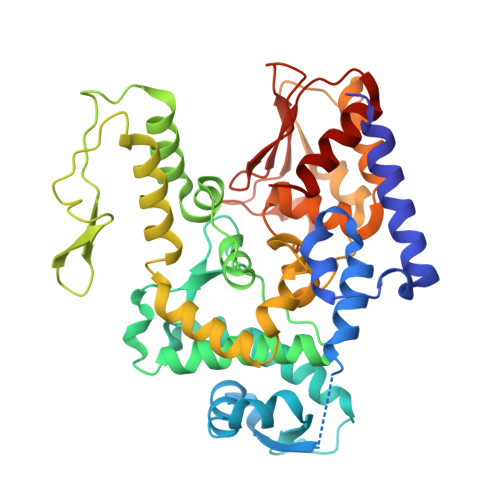
Tom1p ubiquitin ligase structure, interaction with Spt6p, and function in maintaining normal transcript levels and the stability of chromatin in promoters
Madrigal, J., Schubert, H.L., Sdano, M.A., McCullough, L., Connell, Z., Formosa, T., Hill, C.P.(2024) Elife