Structural basis for DCAF2 as a novel E3 ligase for PROTAC-mediated targeted protein degradation.
McMahon, E.J., Cioffi, A.G., Visperas, P.R., Lin, Y., Shaghafi, M., Daczkowski, C.M., Hermann, J.C., Everley, R.A., Neve, R.M., Erlanson, D.A., Webster, K.R., Narayan, V., Wang, W.(2025) Structure 33: 2020-2028.e7
- PubMed: 41045927
- DOI: https://doi.org/10.1016/j.str.2025.09.006
- Primary Citation of Related Structures:
9C5T, 9C5U, 9C5V - PubMed Abstract:
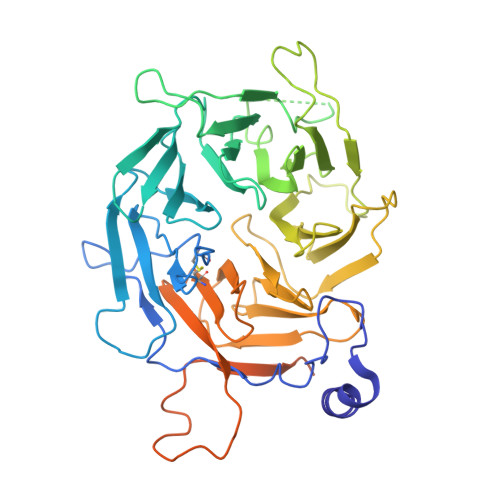
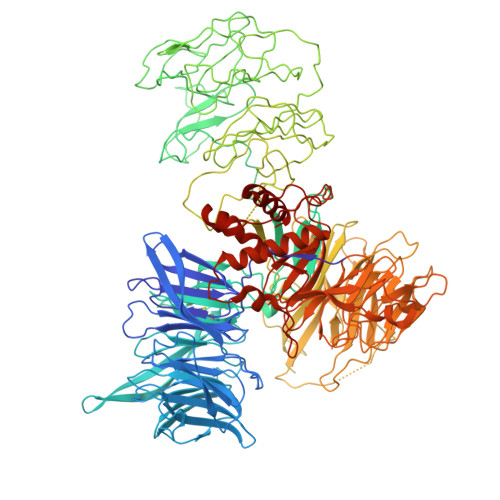
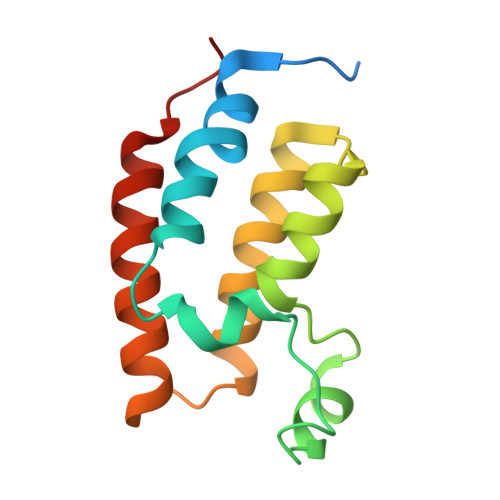
Targeted protein degradation (TPD) leverages the ubiquitin-proteasome system to eliminate disease-causing proteins via E3 ligases. To date, the field is limited to utilizing a few of the over 600 human E3 ligases. To expand this repertoire, we conducted structural and functional validation of DDB1 (Damage-specific DNA binding protein 1) and Cullin-associated factor (DCAF)2 (DTL/CDT2), a Cullin4-RING ligase substrate adaptor implicated in DNA damage response and cancer, as a novel E3 for TPD. Cryoelectron microscopy (cryo-EM) structures of the DCAF2:DDB1:DDA1 complex (3.3 Å), a ligand bound complex (3.1 Å), and a ternary complex with a covalent proteolysis-targeting chimera (PROTAC) and BRD4 (3.4 Å) reveal PROTAC-mediated substrate recruitment. Using covalent bifunctional tool compounds engaging residue C141 in the WD40 domain, we demonstrate robust ubiquitination in biochemical assays and cellular TPD using the COFFEE (covalent functionalization followed by E3 electroporation) method. These findings position DCAF2 as a promising E3 adaptor for PROTAC strategies and identify C141 as a relevant site for future PROTAC discovery.
- Frontier Medicines, 151 Oyster Point Boulevard, South San Francisco, CA 94080, USA.
Organizational Affiliation: