Cryo-EM structure of ATP synthase non-stator state
Zhang, Z., Maharjan, R., Tringides, M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase subunit alpha | A, B [auth C], R [auth B] | 550 | Sus scrofa | Mutation(s): 0 | 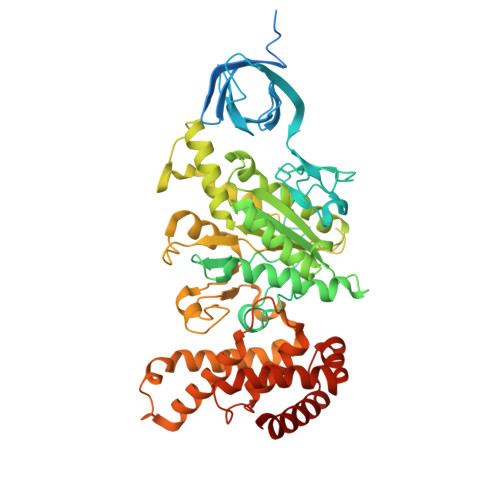 |
UniProt | |||||
Find proteins for A0A8D1XYK3 (Sus scrofa) Explore A0A8D1XYK3 Go to UniProtKB: A0A8D1XYK3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8D1XYK3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase subunit beta | C [auth D], D [auth E], E [auth F] | 570 | Sus scrofa | Mutation(s): 0 EC: 7.1.2.2 | 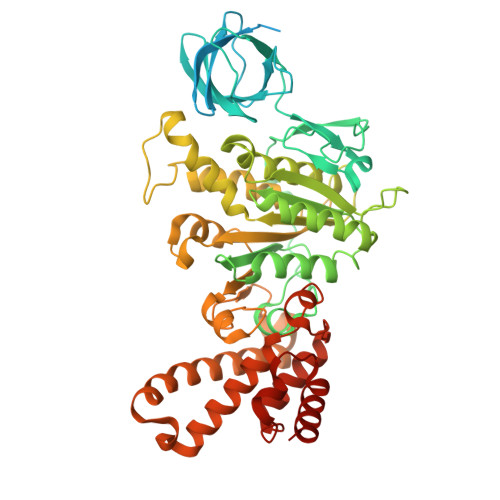 |
UniProt | |||||
Find proteins for A0A8D1JU29 (Sus scrofa) Explore A0A8D1JU29 Go to UniProtKB: A0A8D1JU29 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8D1JU29 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATPase inhibitor, mitochondrial | F [auth J] | 108 | Sus scrofa | Mutation(s): 0 | 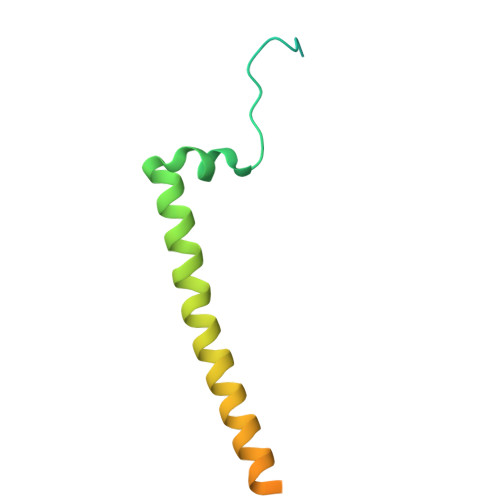 |
UniProt | |||||
Find proteins for Q29307 (Sus scrofa) Explore Q29307 Go to UniProtKB: Q29307 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q29307 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase subunit gamma | 273 | Sus scrofa | Mutation(s): 0 | 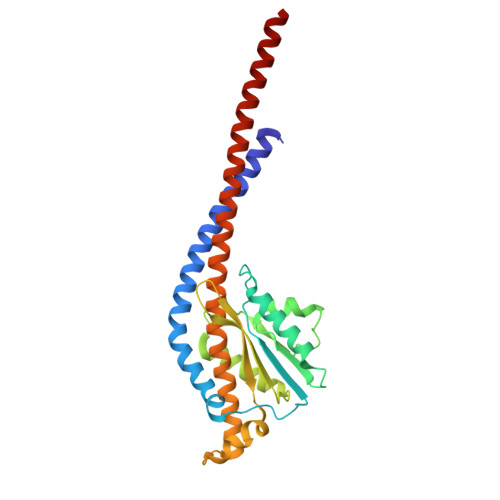 | |
UniProt | |||||
Find proteins for A0A8D0YCC0 (Sus scrofa) Explore A0A8D0YCC0 Go to UniProtKB: A0A8D0YCC0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8D0YCC0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase F1 subunit delta | 168 | Sus scrofa | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A4X1VPE5 (Sus scrofa) Explore A0A4X1VPE5 Go to UniProtKB: A0A4X1VPE5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A4X1VPE5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase F1 subunit epsilon | 136 | Sus scrofa | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A8D1L782 (Sus scrofa) Explore A0A8D1L782 Go to UniProtKB: A0A8D1L782 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8D1L782 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase lipid-binding protein | 141 | Sus scrofa | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A8D1WGE8 (Sus scrofa) Explore A0A8D1WGE8 Go to UniProtKB: A0A8D1WGE8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8D1WGE8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP Query on ATP | AA [auth B], S [auth A], U [auth C] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| ADP Query on ADP | W [auth D], Y [auth F] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| MG Query on MG | T [auth A], V [auth C], X [auth D], Z [auth F] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20.1_4487: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | -- |