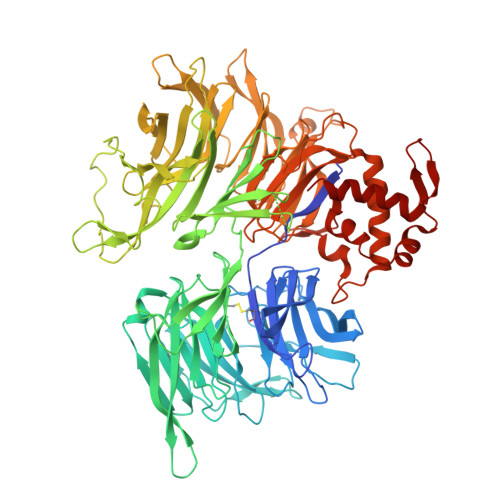
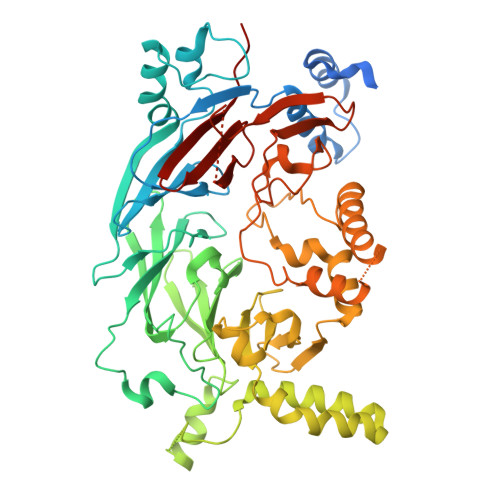
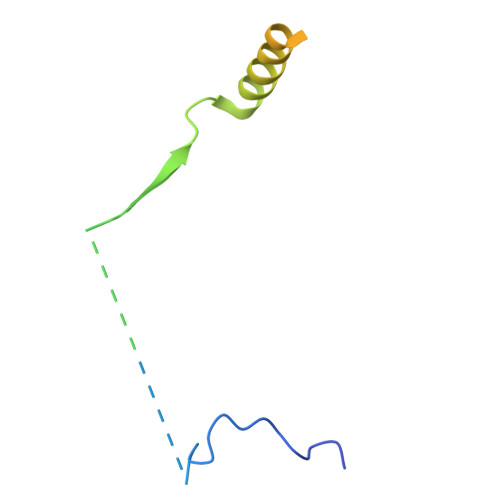
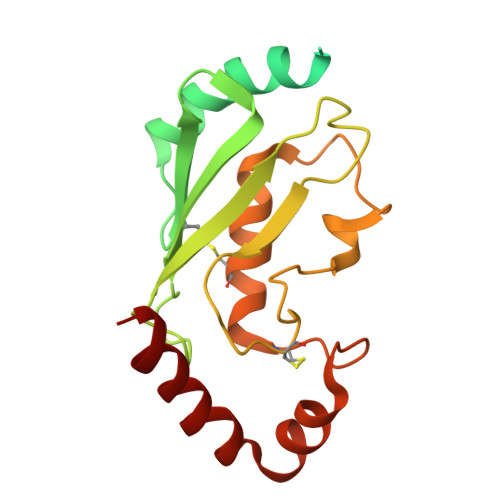
DET1 dynamics underlie cooperative ubiquitination by CRL4 DET1-COP1 complexes.
Burgess, A.E., Loughran, T.A., Turk, L.S., Nyvall, H.G., Dunlop, J.L., Jamieson, S.A., Curry, J.R., Burke, J.E., Filipcik, P., Brown, S.H.J., Mace, P.D.(2025) Sci Adv 11: eadq4187-eadq4187
- PubMed: 40009677
- DOI: https://doi.org/10.1126/sciadv.adq4187
- Primary Citation of Related Structures:
9BJZ - PubMed Abstract:
Transcription factor ubiquitination is a decisive regulator of growth and development. The DET1-DDB1-DDA1 (DDD) complex associates with the Cullin-4 ubiquitin ligase (CRL4) and a second ubiquitin ligase, COP1, to control ubiquitination of transcription factors involved in neurological, metabolic, and immune cell development. Here, we report the structure of the human DDD complex, revealing a specific segment of DET1 that can recruit ubiquitin-conjugating (E2) enzymes. Structural variability analysis, mass spectrometry, and mutagenesis based on AlphaFold predictions suggest that dynamic closure of DET1, stabilized by DDA1, underlies coordinated recruitment of E2 enzymes and COP1. Biochemical assays suggest that the E2 acts as a recruitment factor to bring COP1 to DET1 for more effective substrate ubiquitination, which parallels a catalytically inactive E2 enzyme (COP10) in plant DDD complexes. This work provides a clear architecture for regulation and cooperative CRL4 DET1-COP1 complex assembly, which can affect degradation of diverse targets by COP1 complexes.
- Department of Biochemistry, School of Biomedical Sciences, University of Otago, Dunedin, New Zealand.
Organizational Affiliation: