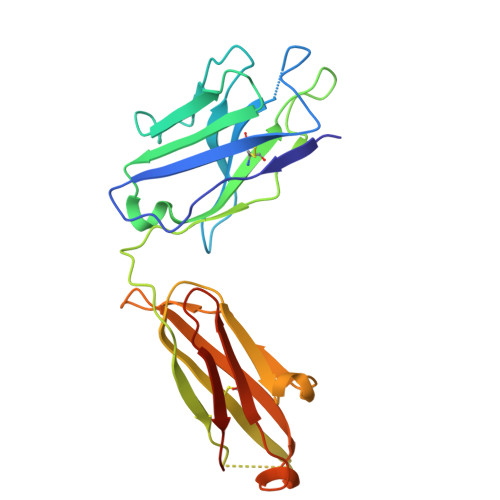
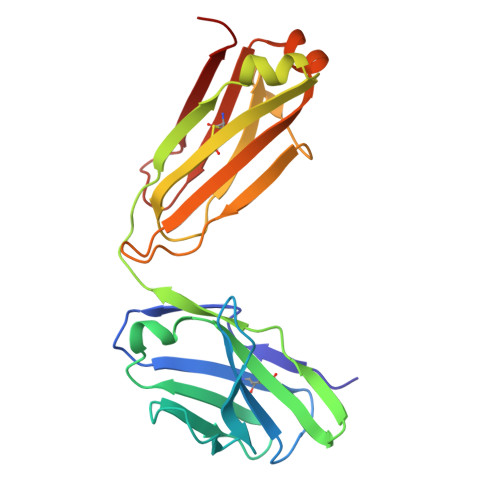
Structure of a Human Monoclonal Antibody in Complex with Outer Surface Protein C of the Lyme Disease Spirochete, Borreliella burgdorferi.
Rudolph, M.J., Chen, Y., Vorauer, C., Vance, D.J., Piazza, C.L., Willsey, G.G., McCarthy, K., Muriuki, B., Cavacini, L.A., Guttman, M., Mantis, N.J.(2024) J Immunol 213: 1234-1243
- PubMed: 39240158
- DOI: https://doi.org/10.4049/jimmunol.2400247
- Primary Citation of Related Structures:
9BIF - PubMed Abstract:
Lyme disease is a tick-borne, multisystem infection caused by the spirochete Borreliella burgdorferi. Although Abs have been implicated in the resolution of Lyme disease, the specific B cell epitopes targeted during human infections remain largely unknown. In this study, we characterized and defined the structural epitope of a patient-derived bactericidal monoclonal IgG (B11) against outer surface protein C (OspC), a homodimeric lipoprotein necessary for B. burgdorferi tick-mediated transmission and early-stage colonization of vertebrate hosts. High-resolution epitope mapping was accomplished through hydrogen deuterium exchange-mass spectrometry and X-ray crystallography. Structural analysis of B11 Fab-OspCA complexes revealed the B11 Fabs associated in a 1:1 stoichiometry with the lateral faces of OspCA homodimers such that the Abs are essentially positioned perpendicular to the spirochete's outer surface. B11's primary contacts reside within the membrane-proximal regions of α-helices 1 and 6 and adjacent loops 5 and 6 in one OspCA monomer. In addition, B11 spans the OspCA dimer interface, engaging opposing α-helix 1', α-helix 2', and loop 2-3' in the second OspCA monomer. The B11-OspCA structure is reminiscent of the recently solved mouse transmission blocking monoclonal IgG B5 in complex with OspCA, indicating a mode of engagement with OspC that is conserved across species. In conclusion, we provide a detailed insight into the interaction between a functional human Ab and an immunodominant Lyme disease Ag long considered an important vaccine candidate.
- New York Structural Biology Center, New York, NY.
Organizational Affiliation: