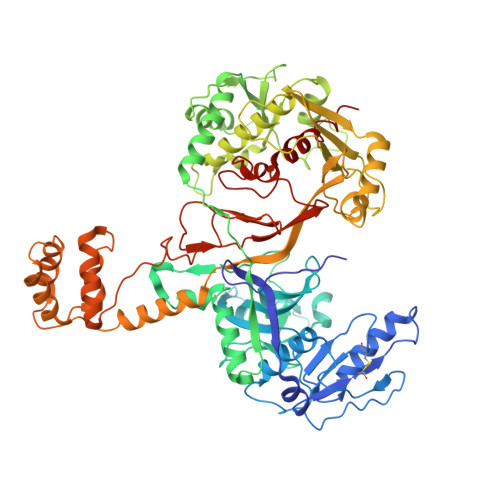
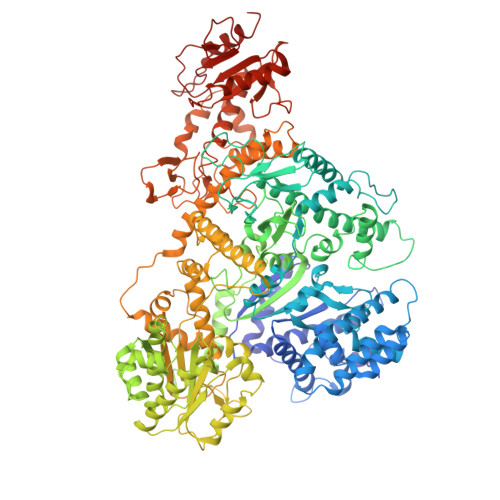
DdmDE eliminates plasmid invasion by DNA-guided DNA targeting.
Yang, X.Y., Shen, Z., Wang, C., Nakanishi, K., Fu, T.M.(2024) Cell 187: 5253-5266.e16
- PubMed: 39173632
- DOI: https://doi.org/10.1016/j.cell.2024.07.028
- Primary Citation of Related Structures:
9BF1, 9BF5, 9BGK, 9C6Q - PubMed Abstract:
Horizontal gene transfer is a key driver of bacterial evolution, but it also presents severe risks to bacteria by introducing invasive mobile genetic elements. To counter these threats, bacteria have developed various defense systems, including prokaryotic Argonautes (pAgos) and the DNA defense module DdmDE system. Through biochemical analysis, structural determination, and in vivo plasmid clearance assays, we elucidate the assembly and activation mechanisms of DdmDE, which eliminates small, multicopy plasmids. We demonstrate that DdmE, a pAgo-like protein, acts as a catalytically inactive, DNA-guided, DNA-targeting defense module. In the presence of guide DNA, DdmE targets plasmids and recruits a dimeric DdmD, which contains nuclease and helicase domains. Upon binding to DNA substrates, DdmD transitions from an autoinhibited dimer to an active monomer, which then translocates along and cleaves the plasmids. Together, our findings reveal the intricate mechanisms underlying DdmDE-mediated plasmid clearance, offering fundamental insights into bacterial defense systems against plasmid invasions.
- Department of Biological Chemistry and Pharmacology, The Ohio State University, Columbus, OH 43210, USA; The Ohio State University Comprehensive Cancer Center, Columbus, OH 43210, USA; Ohio State Biochemistry Program, The Ohio State University, Columbus, OH 43210, USA; Center for RNA Biology, The Ohio State University, Columbus, OH 43210, USA.
Organizational Affiliation: