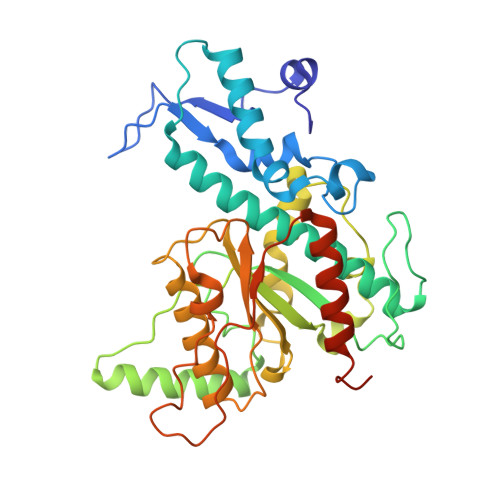
Two-metal ion mechanism of DNA cleavage by activated, filamentous SgrAI.
Shan, Z., Rivero-Gamez, A., Lyumkis, D., Horton, N.C.(2024) J Biological Chem 300: 107576-107576
- PubMed: 39009341
- DOI: https://doi.org/10.1016/j.jbc.2024.107576
- Primary Citation of Related Structures:
9BGI, 9BGJ - PubMed Abstract:
Enzymes that form filamentous assemblies with modulated enzymatic activities have gained increasing attention in recent years. SgrAI is a sequence specific type II restriction endonuclease that forms polymeric filaments with accelerated DNA cleavage activity and expanded DNA sequence specificity. Prior studies have suggested a mechanistic model linking the structural changes accompanying SgrAI filamentation to its accelerated DNA cleavage activity. In this model, the conformational changes that are specific to filamentous SgrAI maximize contacts between different copies of the enzyme within the filament and create a second divalent cation binding site in each subunit, which in turn facilitates the DNA cleavage reaction. However, our understanding of the atomic mechanism of catalysis is incomplete. Herein, we present two new structures of filamentous SgrAI solved using cryo-EM. The first structure, resolved to 3.3 Å, is of filamentous SgrAI containing an active site mutation that is designed to stall the DNA cleavage reaction, which reveals the enzymatic configuration prior to DNA cleavage. The second structure, resolved to 3.1 Å, is of WT filamentous SgrAI containing cleaved substrate DNA, which reveals the enzymatic configuration at the end of the enzymatic cleavage reaction. Both structures contain the phosphate moiety at the cleavage site and the biologically relevant divalent cation cofactor Mg 2+ and define how the Mg 2+ cation reconfigures during enzymatic catalysis. The data support a model for the activation mechanism that involves binding of a second Mg 2+ in the SgrAI active site as a direct result of filamentation induced conformational changes.
- The Salk Institute of Biological Sciences, La Jolla, California, USA.
Organizational Affiliation: