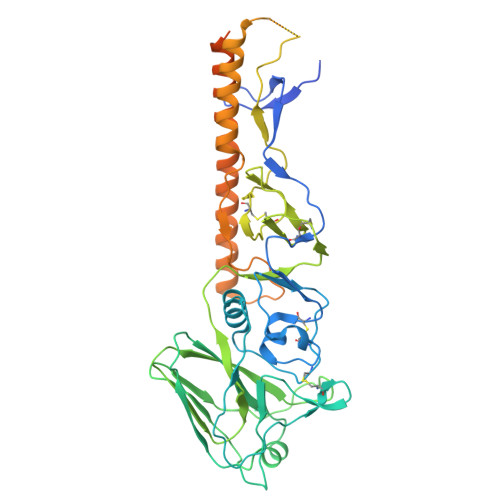
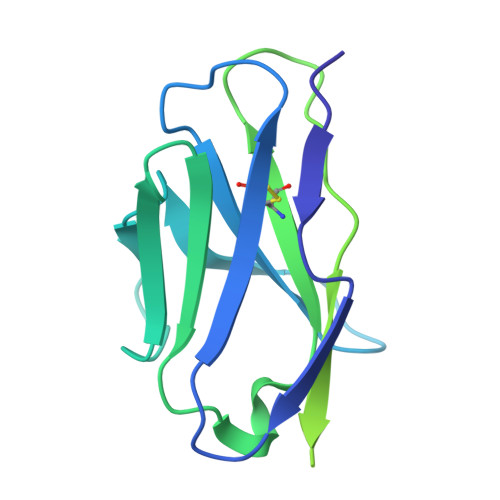
Conserved sites on the influenza H1 and H3 hemagglutinin recognized by human antibodies.
Maurer, D.P., Vu, M., Ferreira Ramos, A.S., Dugan, H.L., Khalife, P., Geoghegan, J.C., Walker, L.M., Bajic, G., Schmidt, A.G.(2025) Sci Adv 11: eadu9140-eadu9140
- PubMed: 40267182
- DOI: https://doi.org/10.1126/sciadv.adu9140
- Primary Citation of Related Structures:
9BDF, 9BDG - PubMed Abstract:
Monoclonal antibodies (mAbs) targeting the influenza hemagglutinin (HA) can be used as prophylactics or templates for next-generation vaccines. Here, we isolated broad, subtype-neutralizing mAbs from human B cells recognizing the H1 or H3 HA "head" and a mAb engaging the conserved stem. The H1 mAbs bind the lateral patch epitope on HAs from 1933 to 2021 and a prepandemic swine H1N1 virus. We improved neutralization potency using directed evolution toward a contemporary H1 HA. Deep mutational scanning of four antigenically distinct H1N1 viruses identified potential viral escape pathways. For the H3 mAbs, we used cryo-electron microscopy to define their epitopes: One mAb binds the side of the HA head, accommodating the N133 glycan and a pocket underneath the receptor binding site; the other mAb recognizes an HA stem epitope that partially overlaps with previously characterized mAbs but with distinct antibody variable genes. Collectively, these mAbs identify conserved sites recognized by broadly-reactive mAbs that may be elicited by next-generation vaccines.
- Ragon Institute of Mass General, MIT, and Harvard, Cambridge, MA 02139, USA.
Organizational Affiliation: