Klebsiella pneumoniae Serine Hydrolase
Randall, G.T.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Esterase | 242 | Klebsiella pneumoniae | Mutation(s): 0 Gene Names: CK244_008325, NCTC13465_04963, NCTC5052_05330, NCTC9601_05351, NCTC9617_04952, NCTC9645_01577, NCTC9661_05640, SAMEA3512100_04172, SAMEA4873632_04766 EC: 3.1.1.23 | 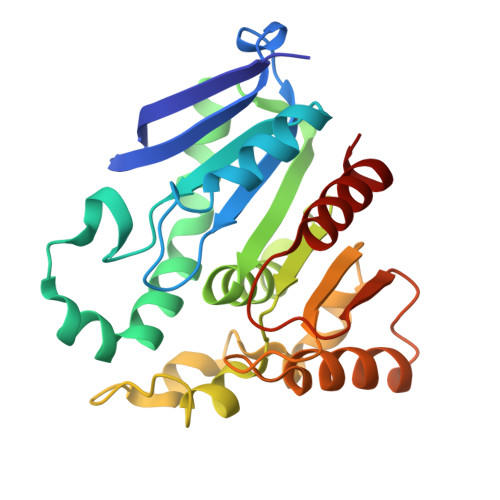 | |
UniProt | |||||
Find proteins for A0A060VRI4 (Klebsiella pneumoniae) Explore A0A060VRI4 Go to UniProtKB: A0A060VRI4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A060VRI4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 102.706 | α = 90 |
| b = 68.195 | β = 77.16 |
| c = 65.363 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| MBIE China-MWC Collaboration | New Zealand | Nr.3720870 |