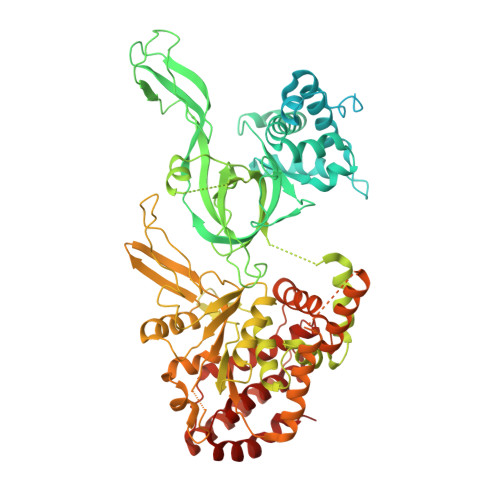
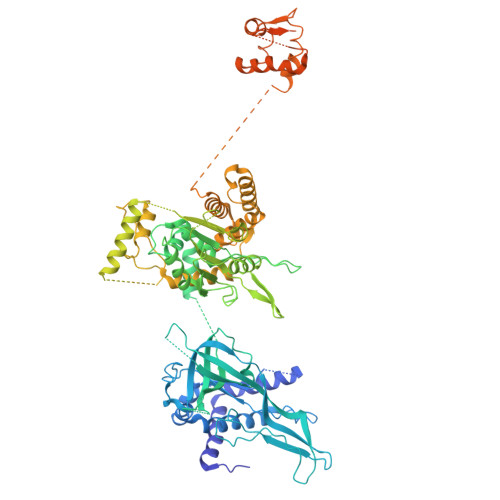
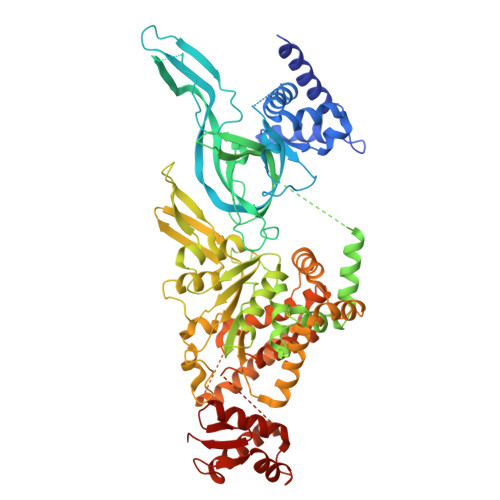
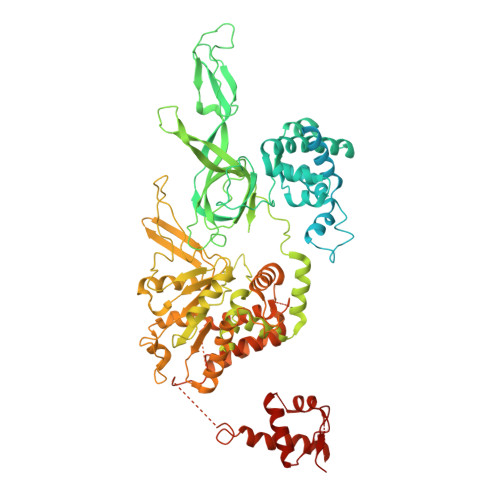
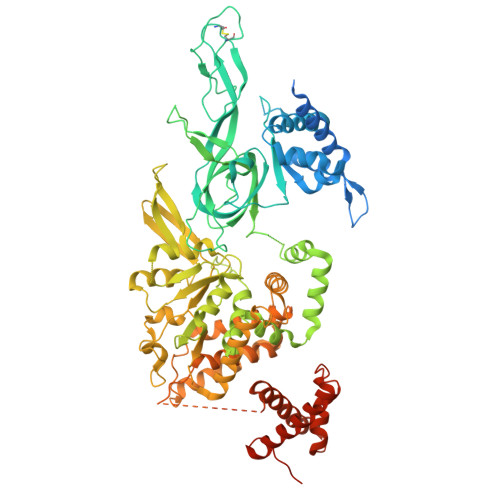
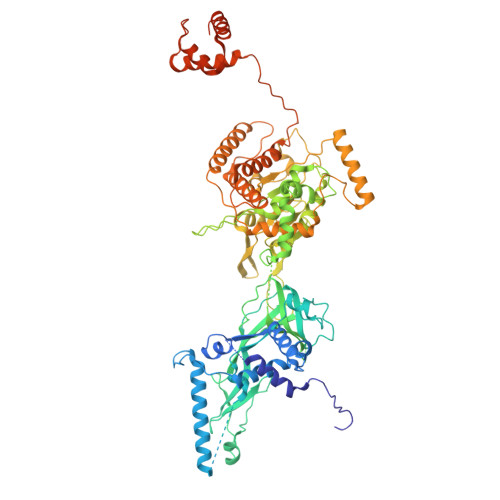
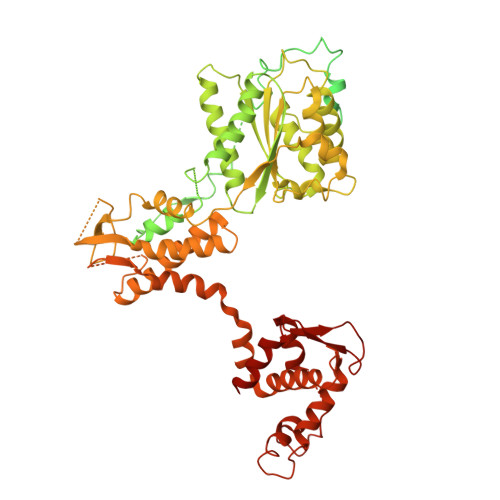
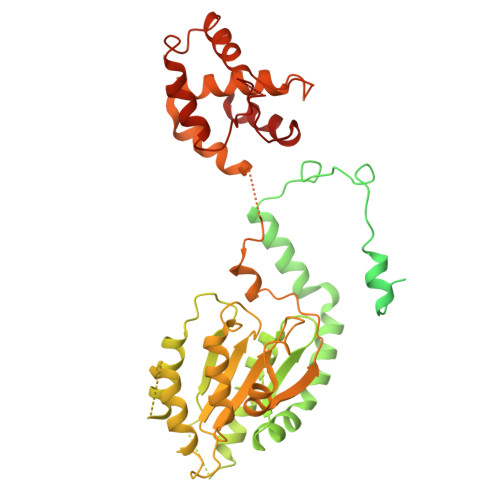
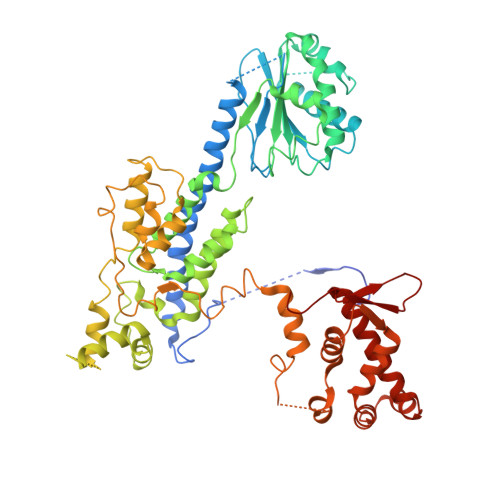
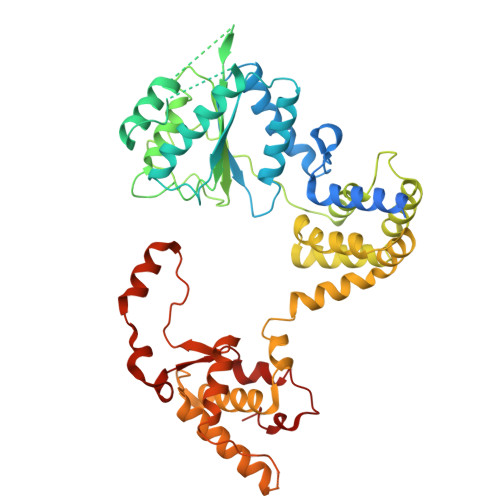
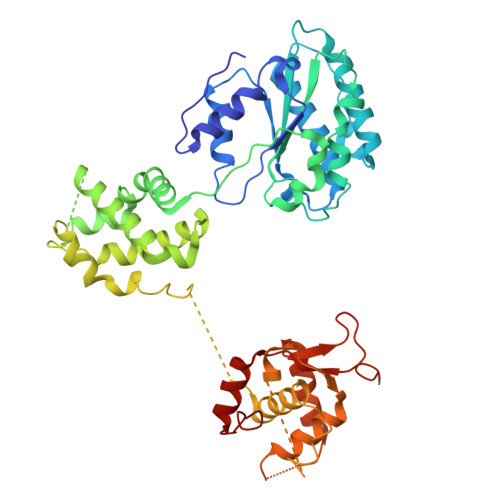
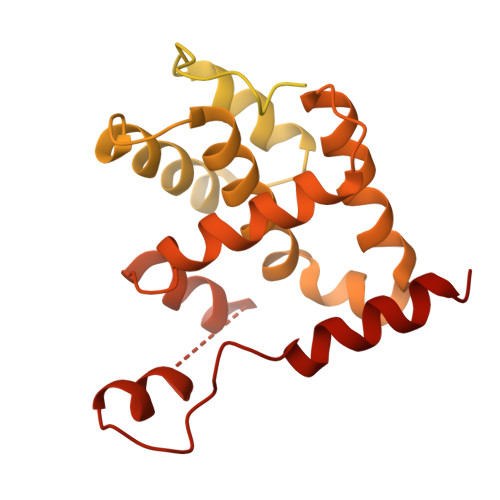
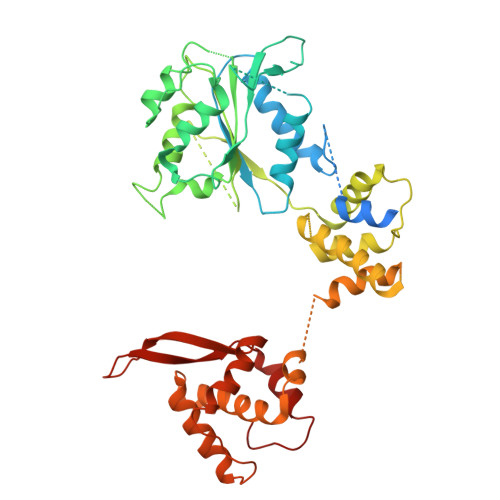
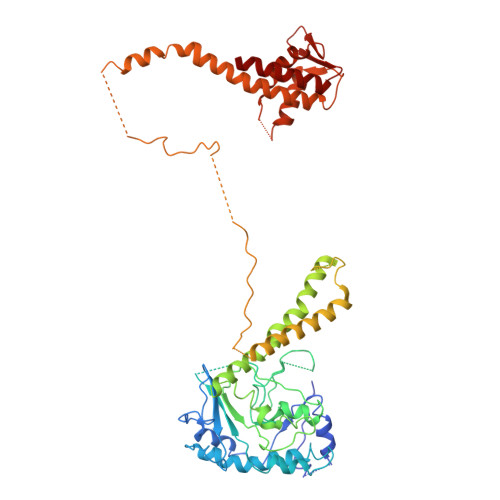
MCM2-7 ring closure involves the Mcm5 C-terminus and triggers Mcm4 ATP hydrolysis.
Faull, S.V., Barbon, M., Mossler, A., Yuan, Z., Bai, L., Reuter, L.M., Riera, A., Winkler, C., Magdalou, I., Peach, M., Li, H., Speck, C.(2025) Nat Commun 16: 14-14
- PubMed: 39747125
- DOI: https://doi.org/10.1038/s41467-024-55479-1
- Primary Citation of Related Structures:
9BCX - PubMed Abstract:
The eukaryotic helicase MCM2-7, is loaded by ORC, Cdc6 and Cdt1 as a double-hexamer onto replication origins. The insertion of DNA into the helicase leads to partial MCM2-7 ring closure, while ATP hydrolysis is essential for consecutive steps in pre-replicative complex (pre-RC) assembly. Currently it is unknown how MCM2-7 ring closure and ATP-hydrolysis are controlled. A cryo-EM structure of an ORC-Cdc6-Cdt1-MCM2-7 intermediate shows a remodelled, fully-closed Mcm2/Mcm5 interface. The Mcm5 C-terminus (C5) contacts Orc3 and specifically recognises this closed ring. Interestingly, we found that normal helicase loading triggers Mcm4 ATP-hydrolysis, which in turn leads to reorganisation of the MCM2-7 complex and Cdt1 release. However, defective MCM2-7 ring closure, due to mutations at the Mcm2/Mcm5 interface, leads to MCM2-7 ring splitting and complex disassembly. As such we identify Mcm4 as the key ATPase in regulating pre-RC formation. Crucially, a stable Mcm2/Mcm5 interface is essential for productive ATP-hydrolysis-dependent remodelling of the helicase.
- DNA Replication Group, Institute of Clinical Science, Imperial College London, London, UK.
Organizational Affiliation: