Structure-guided discovery of viral proteins that inhibit host immunity.
Yirmiya, E., Hobbs, S.J., Leavitt, A., Osterman, I., Avraham, C., Hochhauser, D., Madhala, B., Skovorodka, M., Tan, J.M.J., Toyoda, H.C., Chebotar, I., Itkin, M., Malitsky, S., Amitai, G., Kranzusch, P.J., Sorek, R.(2025) Cell 188: 1681
- PubMed: 39855193
- DOI: https://doi.org/10.1016/j.cell.2024.12.035
- Primary Citation of Related Structures:
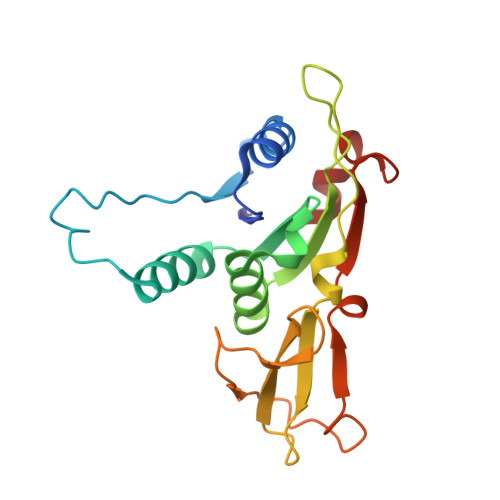
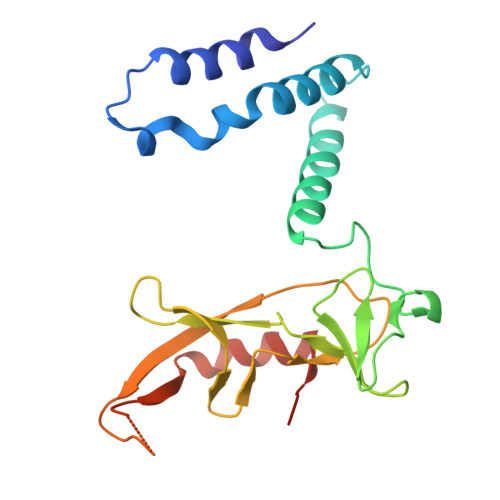
9B7D - PubMed Abstract:
Viruses encode proteins that inhibit host defenses, but sifting through the millions of available viral sequences for immune-modulatory proteins has been so far impractical. Here, we develop a process to systematically screen virus-encoded proteins for inhibitors that physically bind host immune proteins. Focusing on Thoeris and CBASS, bacterial defense systems that are the ancestors of eukaryotic Toll/interleukin-1 receptor (TIR) and cyclic GMP-AMP synthase (cGAS) immunity, we discover seven families of Thoeris and CBASS inhibitors, encompassing thousands of genes widespread in phages. Verified inhibitors exhibit extensive physical interactions with the respective immune protein counterpart, with all inhibitors blocking the active site of the immune protein. Remarkably, a phage-encoded inhibitor of bacterial TIR proteins can bind and inhibit distantly related human and plant immune TIRs, and a phage-derived inhibitor of bacterial cGAS-like enzymes can inhibit the human cGAS. Our results demonstrate that phages are a reservoir for immune-modulatory proteins capable of inhibiting bacterial, animal, and plant immunity.
- Department of Molecular Genetics, Weizmann Institute of Science, Rehovot 7610001, Israel.
Organizational Affiliation: