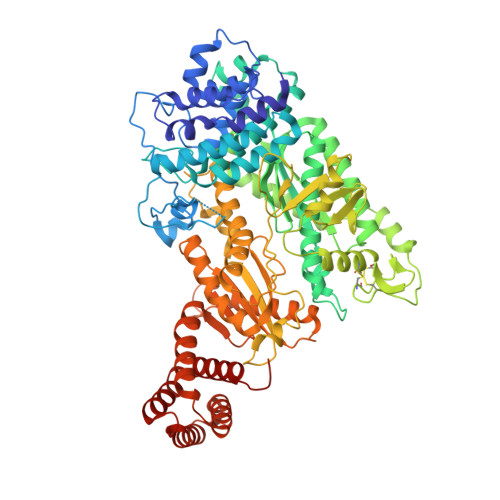
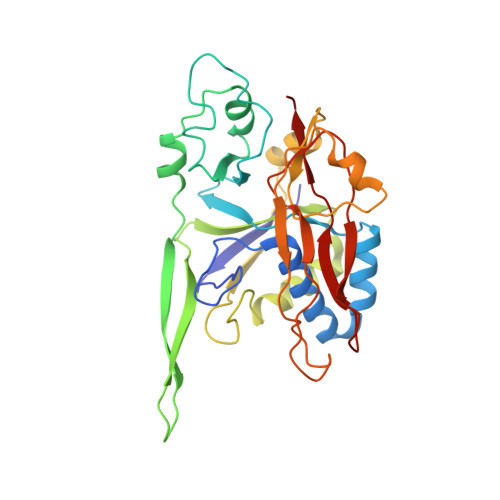
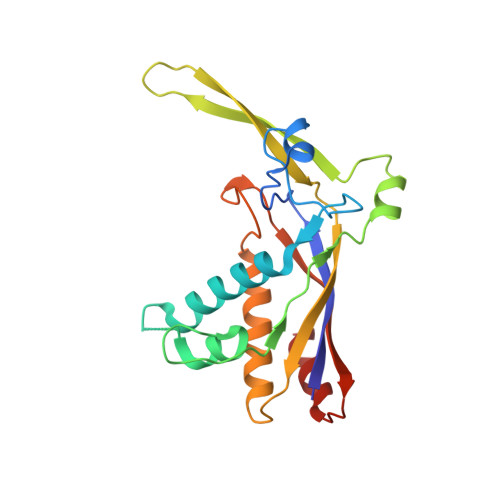
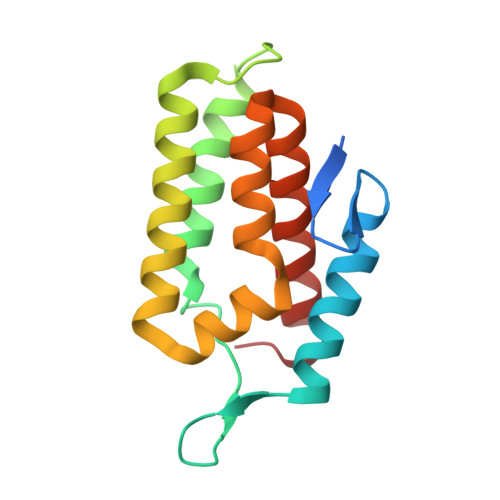
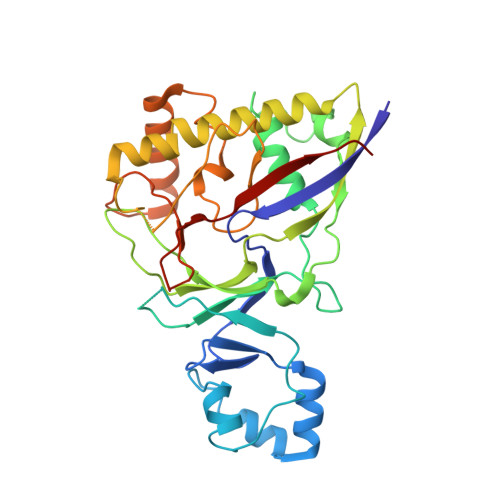
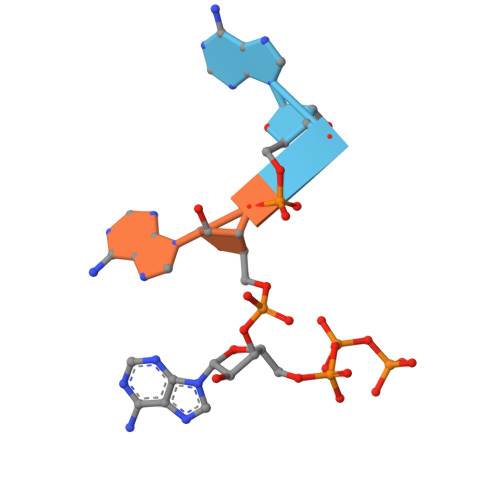
Molecular basis for cA6 synthesis by a type III-A CRISPR-Cas enzyme and its conversion to cA4 production.
Goswami, H.N., Ahmadizadeh, F., Wang, B., Addo-Yobo, D., Zhao, Y., Whittington, A.C., He, H., Terns, M.P., Li, H.(2024) Nucleic Acids Res 52: 10619-10629
- PubMed: 38989619
- DOI: https://doi.org/10.1093/nar/gkae603
- Primary Citation of Related Structures:
9ASH, 9ASI - PubMed Abstract:
The type III-A (Csm) CRISPR-Cas systems are multi-subunit and multipronged prokaryotic enzymes in guarding the hosts against viral invaders. Beyond cleaving activator RNA transcripts, Csm confers two additional activities: shredding single-stranded DNA and synthesizing cyclic oligoadenylates (cOAs) by the Cas10 subunit. Known Cas10 enzymes exhibit a fascinating diversity in cOA production. Three major forms-cA3, cA4 and cA6have been identified, each with the potential to trigger unique downstream effects. Whereas the mechanism for cOA-dependent activation is well characterized, the molecular basis for synthesizing different cOA isoforms remains unclear. Here, we present structural characterization of a cA6-producing Csm complex during its activation by an activator RNA. Analysis of the captured intermediates of cA6 synthesis suggests a 3'-to-5' nucleotidyl transferring process. Three primary adenine binding sites can be identified along the chain elongation path, including a unique tyrosine-threonine dyad found only in the cA6-producing Cas10. Consistently, disrupting the tyrosine-threonine dyad specifically impaired cA6 production while promoting cA4 production. These findings suggest that Cas10 utilizes a unique enzymatic mechanism for forming the phosphodiester bond and has evolved distinct strategies to regulate the cOA chain length.
- Institute of Molecular Biophysics, Florida State University, Tallahassee, FL 32306, USA.
Organizational Affiliation: