Structure of a type III CRISPR complex that AMPylates a HEPN toxin
Pandey, S., Burman, N., Henriques, W.H., Zahl, T., Wiegand, T., Nyquist, H., Wilkinson, R., Wiedenheft, B.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CSD domain-containing protein Cmr6 | A [auth H] | 376 | Dissulfurispira thermophila | Mutation(s): 0 Gene Names: JZK55_14930 | 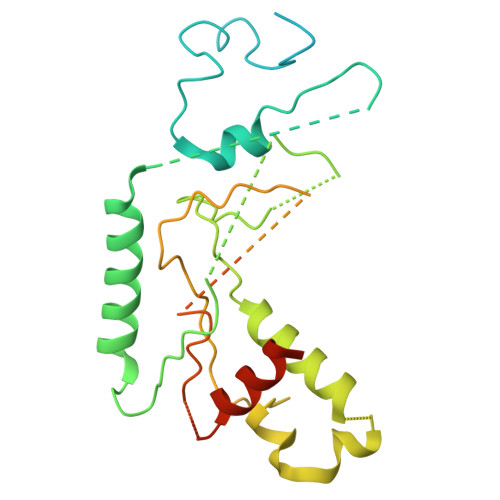 |
UniProt | |||||
Find proteins for A0A7G1H339 (Dissulfurispira thermophila) Explore A0A7G1H339 Go to UniProtKB: A0A7G1H339 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A7G1H339 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Type III-B CRISPR-associated protein Cas10/Cmr2 | B [auth A] | 612 | Dissulfurispira thermophila | Mutation(s): 0 Gene Names: JZK55_14980 | 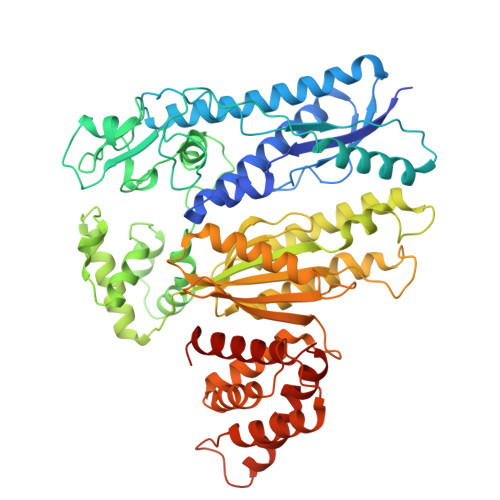 |
UniProt | |||||
Find proteins for A0A7G1H3Q2 (Dissulfurispira thermophila) Explore A0A7G1H3Q2 Go to UniProtKB: A0A7G1H3Q2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A7G1H3Q2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Type III-B CRISPR module RAMP protein Cmr4 | 315 | Dissulfurispira thermophila | Mutation(s): 0 Gene Names: JZK55_14950 | 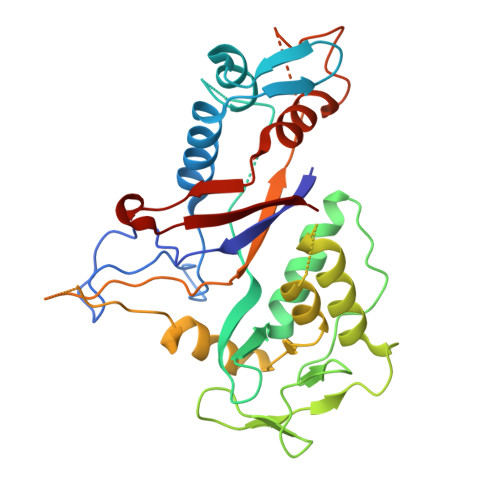 | |
UniProt | |||||
Find proteins for A0A7G1H376 (Dissulfurispira thermophila) Explore A0A7G1H376 Go to UniProtKB: A0A7G1H376 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A7G1H376 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CRISPR type III-B/RAMP module-associated protein Cmr5 | 140 | Dissulfurispira thermophila | Mutation(s): 0 Gene Names: JZK55_14940 | 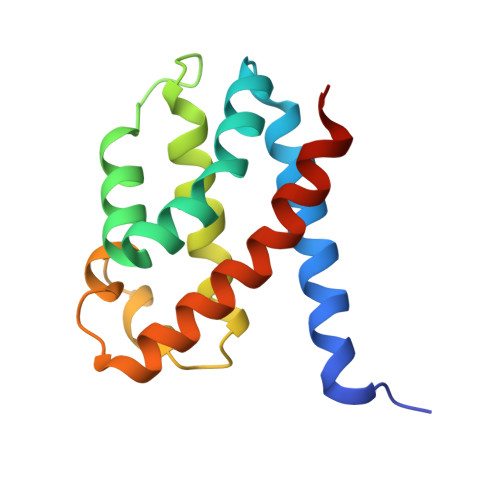 | |
UniProt | |||||
Find proteins for A0A7G1H353 (Dissulfurispira thermophila) Explore A0A7G1H353 Go to UniProtKB: A0A7G1H353 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A7G1H353 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Type III-B CRISPR module-associated protein Cmr3 | H [auth B] | 360 | Dissulfurispira thermophila | Mutation(s): 0 Gene Names: JZK55_14970 | 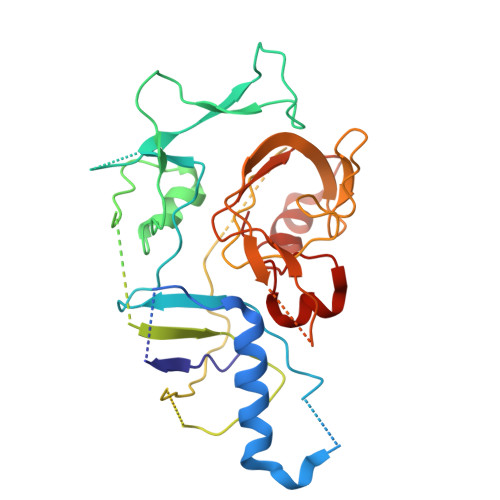 |
UniProt | |||||
Find proteins for A0A7G1H1A4 (Dissulfurispira thermophila) Explore A0A7G1H1A4 Go to UniProtKB: A0A7G1H1A4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A7G1H1A4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN (Subject of Investigation/LOI) Query on ZN | I [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20.1_4487: |
| RECONSTRUCTION | cryoSPARC | 4.1.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R35GM134867-01 |