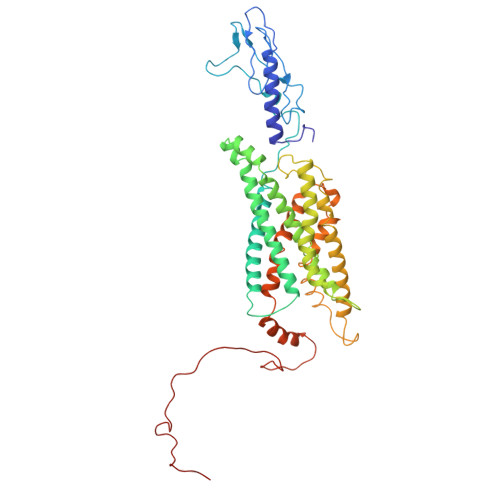
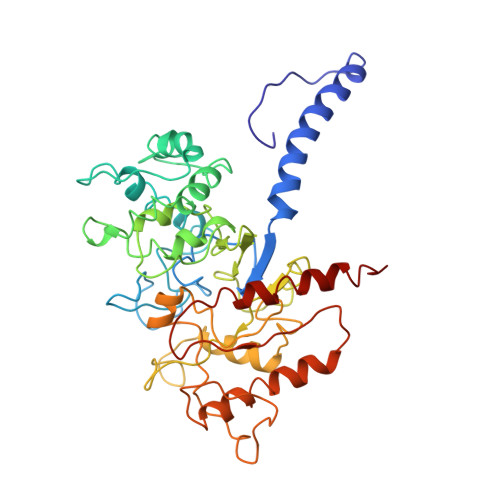
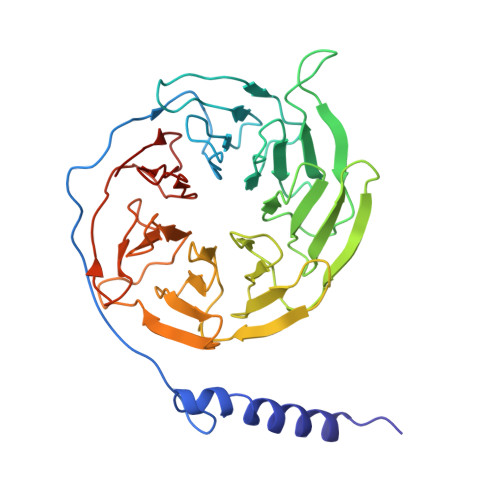
Conformational Dynamics of the Activated GLP-1 Receptor-Gs Complex Revealed by Cross-Linking Mass Spectrometry and Integrative Structure Modeling
Yuan, S., Xia, L., Wang, C., Wu, F., Zhang, B., Pan, C., Fan, Z., Lei, X., Stevens, R., Sali, A., Sun, L., Shui, W.(2023) Acs Cent Sci 9: 992-1007
- PubMed: 37252352
- DOI: https://doi.org/10.1021/acscentsci.3c00063
- Primary Citation of Related Structures:
9A3F - PubMed Abstract:
Despite advances in characterizing the structures and functions of G protein-coupled receptors (GPCRs), our understanding of GPCR activation and signaling is still limited by the lack of information on conformational dynamics. It is particularly challenging to study the dynamics of GPCR complexes with their signaling partners because of their transient nature and low stability. Here, by combining cross-linking mass spectrometry (CLMS) with integrative structure modeling, we map the conformational ensemble of an activated GPCR-G protein complex at near-atomic resolution. The integrative structures describe heterogeneous conformations for a high number of potential alternative active states of the GLP-1 receptor-G s complex. These structures show marked differences from the previously determined cryo-EM structure, especially at the receptor-G s interface and in the interior of the G s heterotrimer. Alanine-scanning mutagenesis coupled with pharmacological assays validates the functional significance of 24 interface residue contacts only observed in the integrative structures, yet absent in the cryo-EM structure. Through the integration of spatial connectivity data from CLMS with structure modeling, our study provides a new approach that is generalizable to characterizing the conformational dynamics of GPCR signaling complexes.
- iHuman Institute, ShanghaiTech University, Shanghai 201210, China.
Organizational Affiliation: