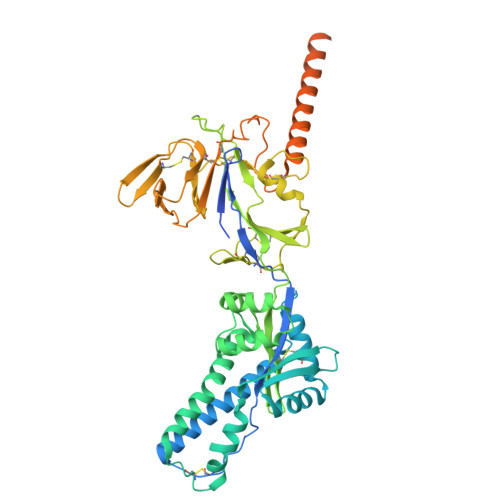
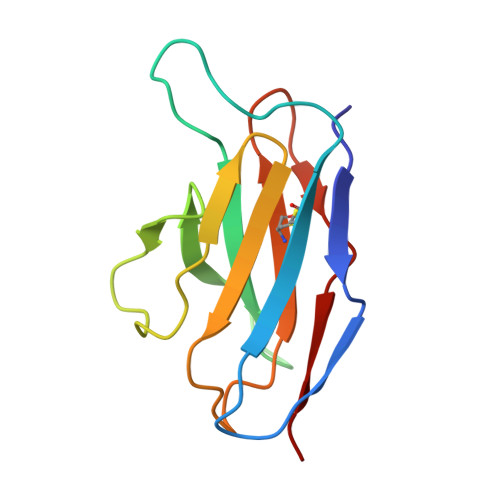
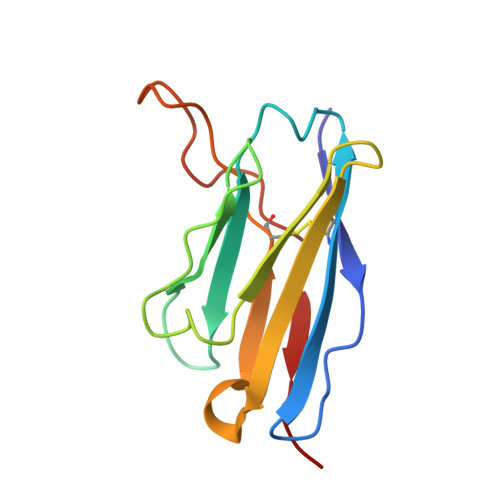
DS2 designer pre-fusion F vaccine induces strong and protective antibody response against RSV infection.
Yang, Y., Wang, R., Guo, F., Zhao, T., Lei, Y., Yang, Q., Zeng, Y., Yang, Z., Ajavavarakula, T., Tan, R., Li, M., Dong, H., Niu, M., Bao, K., Geng, H., Lv, Q., Zhang, Q., Shi, X., Liu, P., Ge, J., Wang, X., Zhang, L.(2024) NPJ Vaccines 9: 258-258
- PubMed: 39741146
- DOI: https://doi.org/10.1038/s41541-024-01059-9
- Primary Citation of Related Structures:
8ZYM - PubMed Abstract:
DS-Cav1, SC-TM, and DS2 are distinct designer pre-fusion F proteins (pre-F) of respiratory syncytial virus (RSV) developed for vaccines. However, their immunogenicity has not been directly compared. In this study, we generated three recombinant vaccines using the chimpanzee adenovirus vector AdC68 to express DS-Cav1, SC-TM, and DS2. All three vaccines elicited robust serum binding and neutralizing antibodies following intramuscular priming and boosting. DS2 induced the strongest antibody responses, followed by SC-TM and DS-Cav1. DS2 also provided strong protection against live RSV challenge. Monoclonal antibodies (mAbs) isolated from long-lived antibody-secreting cells (ASCs) in the bone marrow six months post-immunization with AdC68-DS2 predominantly targeted site Ø as well as site II. One neutralizing antibody against site II, mAb60, conferred strong protection against live RSV infection in mice. These findings highlight the strong ability of the DS2 design in eliciting long-lived antibody responses and guide the development of next-generation RSV vaccines.
- Comprehensive AIDS Research Center, Pandemic Research Alliance Unit, Center for Infection Biology, School of Basic Medical Sciences, Tsinghua University, 100084, Beijing, China.
Organizational Affiliation: