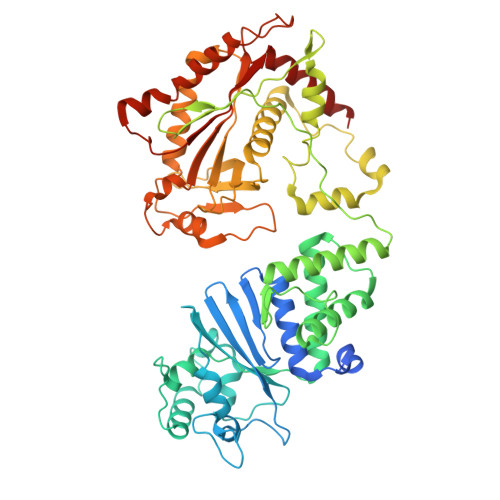
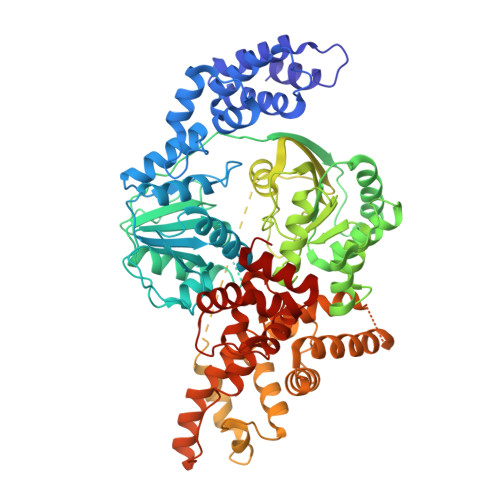
Bacterial Hachiman complex executes DNA cleavage for antiphage defense.
Cui, Y., Dai, Z., Ouyang, Y., Fu, C., Wang, Y., Chen, X., Yang, K., Zheng, S., Wang, W., Tao, P., Guan, Z., Zou, T.(2025) Nat Commun 16: 2604-2604
- PubMed: 40097437
- DOI: https://doi.org/10.1038/s41467-025-57851-1
- Primary Citation of Related Structures:
8ZUE - PubMed Abstract:
Bacteria have developed a variety of immune systems to combat phage infections. The Hachiman system is a novel prokaryotic antiphage defense system comprising HamA and HamB proteins, which contains the DUF1837 and helicase domains, respectively. However, the defense mechanism remains only partially understood. Here, we present the cryo-electron microscopy (cryo-EM) structure of the Hachiman defense system featuring a fusion of Cap4 nuclease domain within HamA. Further structure analysis indicates that the DUF1837 domain on HamA resembles the PD-(D/E)XK nuclease but lacks active sites. Bioinformatics analysis reveals that catalytically inactive DUF1837 domains often recruit other functional domains to fulfill anti-phage defense. HamA interacts with HamB to form a heterodimer HamAB to mediate ATP hydrolysis and execute DNA cleavage, thus implementing antiphage defense. Our findings elucidate the structural basis of the Hachiman defense complex, highlighting the critical roles of the helicase and nuclease in prokaryotic immunity.
- National Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, China.
Organizational Affiliation: