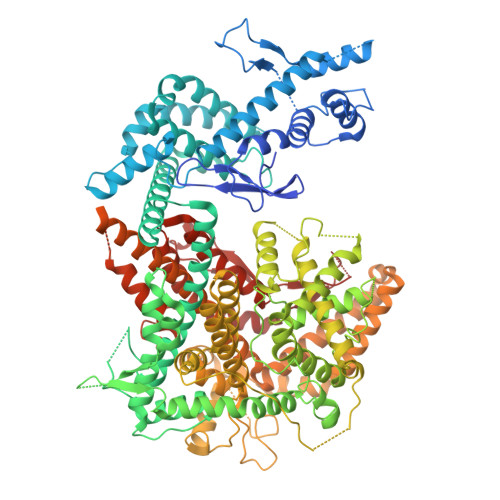
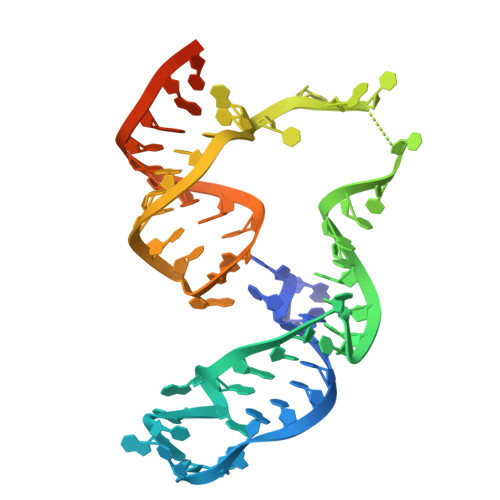
RNA-mediated CRISPR-Cas13 inhibition through crRNA structural mimicry.
Hayes, V.M., Zhang, J.T., Katz, M.A., Li, Y., Kocsis, B., Brinkley, D.M., Jia, N., Meeske, A.J.(2025) Science 388: 387-391
- PubMed: 40273258
- DOI: https://doi.org/10.1126/science.adr3656
- Primary Citation of Related Structures:
8ZTY - PubMed Abstract:
To circumvent CRISPR-Cas immunity, phages express anti-CRISPR factors that inhibit the expression or activities of Cas proteins. Whereas most anti-CRISPRs described to date are proteins, recently described small RNAs called RNA anti-CRISPRs (rAcrs) have sequence homology to CRISPR RNAs (crRNAs) and displace them from cognate Cas nucleases. In this work, we report the discovery of rAcrVIA1-a plasmid-encoded small RNA that inhibits the RNA-targeting CRISPR-Cas13 system in its natural host, Listeria seeligeri . We solved the cryo-electron microscopy structure of the Cas13-rAcr complex, which revealed that rAcrVIA1 adopts a fold nearly identical to crRNA despite sharing negligible sequence similarity. Collectively, our findings expand the diversity of rAcrs and reveal an example of immune antagonism through RNA structural mimicry.
- Department of Microbiology, University of Washington, Seattle, WA, USA.
Organizational Affiliation: