Cryo-EM structure of pyraclostrobin-bound Arachis hypogaea bc1 complex
Cui, G.R., Wang, Y.X., Yang, G.F.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitochondrial-processing peptidase subunit alpha | A, K [auth M] | 460 | Arachis hypogaea | Mutation(s): 0 | 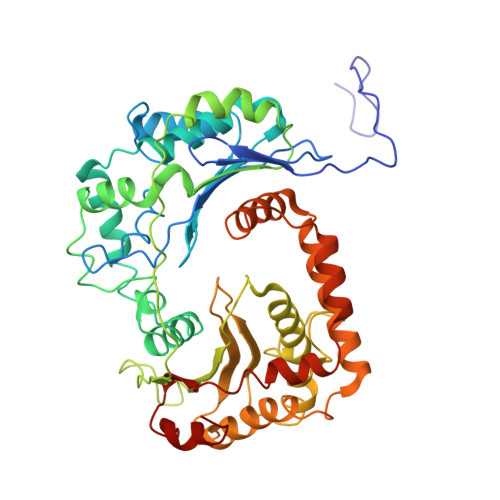 |
UniProt | |||||
Find proteins for A0A445DY18 (Arachis hypogaea) Explore A0A445DY18 Go to UniProtKB: A0A445DY18 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A445DY18 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitochondrial-processing peptidase subunit beta | B, L [auth N] | 487 | Arachis hypogaea | Mutation(s): 0 EC: 3.4.24.64 | 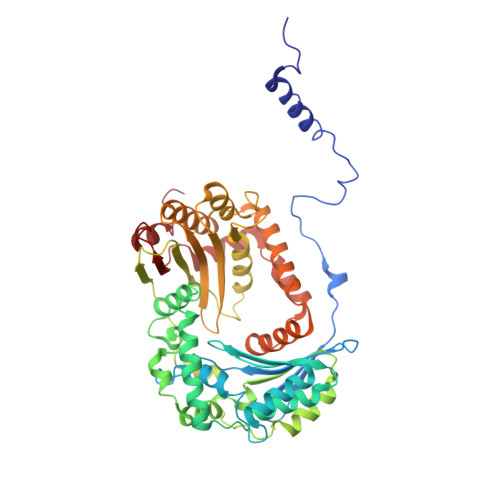 |
UniProt | |||||
Find proteins for A0A445CDV5 (Arachis hypogaea) Explore A0A445CDV5 Go to UniProtKB: A0A445CDV5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A445CDV5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b | C, M [auth O] | 386 | Arachis hypogaea | Mutation(s): 0 | 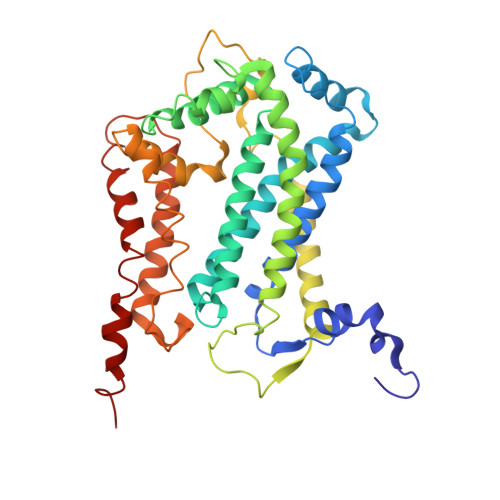 |
UniProt | |||||
Find proteins for A0A8F2YUY6 (Arachis hypogaea) Explore A0A8F2YUY6 Go to UniProtKB: A0A8F2YUY6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8F2YUY6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c domain-containing protein | D, N [auth P] | 242 | Arachis hypogaea | Mutation(s): 0 | 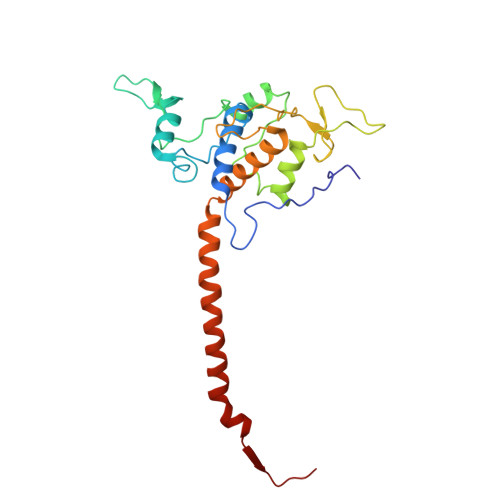 |
UniProt | |||||
Find proteins for A0A445EV57 (Arachis hypogaea) Explore A0A445EV57 Go to UniProtKB: A0A445EV57 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A445EV57 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b-c1 complex subunit Rieske, mitochondrial | E, O [auth Q] | 196 | Arachis hypogaea | Mutation(s): 0 EC: 7.1.1.8 |  |
UniProt | |||||
Find proteins for A0A444YS04 (Arachis hypogaea) Explore A0A444YS04 Go to UniProtKB: A0A444YS04 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A444YS04 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b-c1 complex subunit 7 | F, P [auth R] | 117 | Arachis hypogaea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A445CVZ9 (Arachis hypogaea) Explore A0A445CVZ9 Go to UniProtKB: A0A445CVZ9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A445CVZ9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b-c1 complex subunit 8 | G, Q [auth S] | 70 | Arachis hypogaea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A445EVJ9 (Arachis hypogaea) Explore A0A445EVJ9 Go to UniProtKB: A0A445EVJ9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A445EVJ9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b-c1 complex subunit 6 | H, R [auth T] | 64 | Arachis hypogaea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A444WZ62 (Arachis hypogaea) Explore A0A444WZ62 Go to UniProtKB: A0A444WZ62 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A444WZ62 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Complex III subunit 9 | I [auth J], S [auth V] | 60 | Arachis hypogaea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A445CQN5 (Arachis hypogaea) Explore A0A445CQN5 Go to UniProtKB: A0A445CQN5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A445CQN5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ubiquinol-cytochrome c reductase complex 6.7 kDa protein | J [auth K], T [auth W] | 29 | Arachis hypogaea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A0A6ZDN8 (Arachis hypogaea) Explore A0A0A6ZDN8 Go to UniProtKB: A0A0A6ZDN8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0A6ZDN8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 8 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CDL (Subject of Investigation/LOI) Query on CDL | FA [auth C] LA [auth N] MA [auth N] QA [auth O] TA [auth O] | CARDIOLIPIN C81 H156 O17 P2 XVTUQDWPJJBEHJ-KZCWQMDCSA-L |  | ||
| PC1 (Subject of Investigation/LOI) Query on PC1 | WA [auth Q] | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE C44 H88 N O8 P NRJAVPSFFCBXDT-HUESYALOSA-N |  | ||
| 3PE (Subject of Investigation/LOI) Query on 3PE | AA [auth C] BA [auth C] CA [auth C] DA [auth C] EA [auth C] | 1,2-Distearoyl-sn-glycerophosphoethanolamine C41 H82 N O8 P LVNGJLRDBYCPGB-LDLOPFEMSA-N |  | ||
| HEC (Subject of Investigation/LOI) Query on HEC | HA [auth D], UA [auth P] | HEME C C34 H34 Fe N4 O4 HXQIYSLZKNYNMH-LJNAALQVSA-N |  | ||
| HEM (Subject of Investigation/LOI) Query on HEM | OA [auth O], PA [auth O], X [auth C], Y [auth C] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| A1D6K Query on A1D6K | NA [auth O], W [auth C] | methyl ~{N}-[2-[[1-(4-chlorophenyl)pyrazol-3-yl]oxymethyl]phenyl]-~{N}-methoxy-carbamate C19 H18 Cl N3 O4 HZRSNVGNWUDEFX-UHFFFAOYSA-N |  | ||
| FES (Subject of Investigation/LOI) Query on FES | IA [auth E], VA [auth Q] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | KA [auth N], U [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |