Structural and mechanistic insights into the activation of a short prokaryotic argonaute system from archaeon Sulfolobus islandicus.
Dai, Z., Chen, Y., Guan, Z., Chen, X., Tan, K., Yang, K., Yan, X., Liu, Y., Gong, Z., Han, W., Zou, T.(2025) Nucleic Acids Res 53
- PubMed: 39898546
- DOI: https://doi.org/10.1093/nar/gkaf059
- Primary Citation of Related Structures:
8ZNJ, 9LGW - PubMed Abstract:
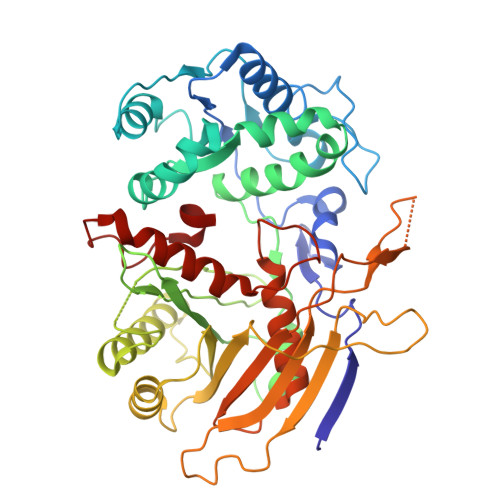
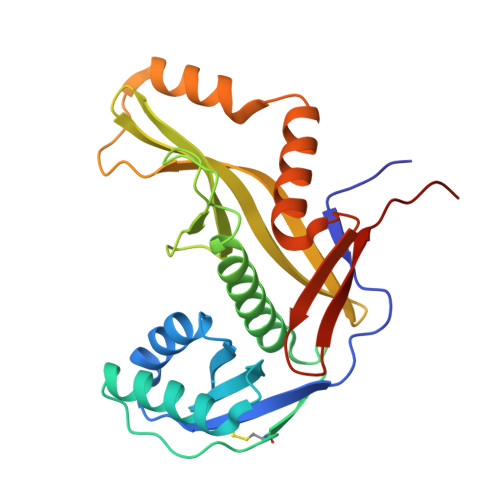
Prokaryotic Argonaute proteins (pAgos) defend the host against invading nucleic acids, including plasmids and viruses. Short pAgo systems confer immunity by inducing cell death upon detecting invading nucleic acids. However, the activation mechanism of the SiAgo system, comprising a short pAgo from the archaeon Sulfolobus islandicus and its associated proteins SiAga1 and SiAga2, remains largely unknown. Here, we determined the cryo-electron microscopy structures of the SiAgo-Aga1 apo complex and the RNA-DNA-bound SiAgo-Aga1 complex at resolutions of 2.7 and 3.0 Å, respectively. Our results revealed that a positively charged pocket is generated from the interaction between SiAgo and SiAga1, exhibiting an architecture similar to APAZ-pAgo of short pAgo systems and accommodating the nucleic acids. Further investigation elucidated the conserved mechanism of nucleic acid recognition by SiAgo-Aga1. Both the SiAgo-Aga1 interaction and nucleic acid recognition by the complex are essential for antiviral defense. Biochemical and structural analyses demonstrated that SiAgo-Aga1 undergoes extensive conformational changes upon binding to the RNA-DNA duplex, thereby licensing its interaction with the effector SiAga2 to trigger the immune response. Overall, our findings highlight the evolutionary conservation of Agos across phylogenetic clades and provide structural insights into the activation mechanism of the SiAgo system.
- National Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, Huazhong Agricultural University, 430070 Wuhan, Hubei, China.
Organizational Affiliation: