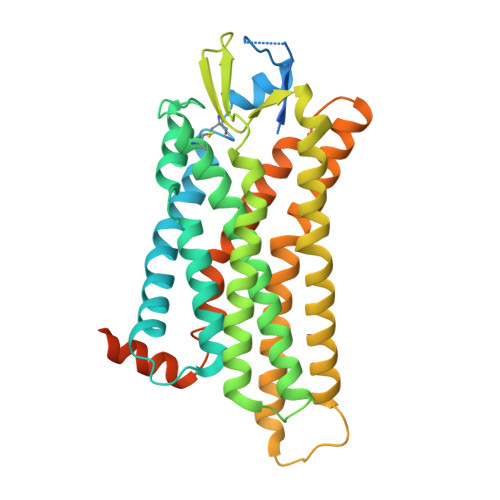
Structural basis of protease-activated receptor 2 activation and biased agonism.
Zhu, X., Xia, R., Zhang, A., Guo, C., Xu, Z., He, Y.(2025) Cell Discov 11: 96-96
- PubMed: 41330898
- DOI: https://doi.org/10.1038/s41421-025-00851-8
- Primary Citation of Related Structures:
8ZMD, 8ZME - PubMed Abstract:
Protease-activated receptor 2 (PAR2) is a transmembrane receptor that is irreversibly activated by proteolytic cleavage of its N-terminus via extracellular proteases, resulting in the release of the tethered ligand (TL), which binds to and activates the receptor. PAR2 plays a pivotal role in the inflammatory response and pain sensation and is a promising drug target for treating arthritis, asthma, and neuronal pain. Here, we present the cryo-electron microscopy structures of active PAR2 complexed with miniG s/q and miniG 13 . Combining functional assays with structural analysis, our study revealed that TL forms a parallel β-sheet with the extracellular loop 2 of PAR2 to engage the receptor. The binding of TL triggers a conformational rearrangement in the transmembrane core, releasing the inhibitory ion lock and allowing receptor activation. Furthermore, we provide structural insights into the engagement of G q and G 13 with PAR2, highlighting that a hydrophobic interaction mediated by the last methionine residue of Gα 13 is crucial for G 13 coupling selectivity. In combination with molecular dynamics simulations and mutagenesis, we identified the I39 TL3 /D62 N-term interaction at the pocket side of the receptor as a key determinant of G 13 signaling. Disrupting this interaction significantly inhibits G 13 signaling while preserving G q activity, enabling us to design a biased peptide ligand that selectively activates G q signaling. The information revealed in this study provides a framework for understanding PAR2 signaling and offers a rational basis for the design of biased PAR2 ligands.
- HIT Center for Life Sciences, School of Life Science and Technology, Faculty of Life Sciences and Medicine, Harbin Institute of Technology, Harbin, Heilongjiang, China.
Organizational Affiliation: