membrane transporter
Zhang, B., Xiong, Z., Rao, Z.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Siderophore exporter MmpL5 | A, D [auth B], G [auth E] | 964 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: mmpL5, Rv0676c, MTV040.04c Membrane Entity: Yes | 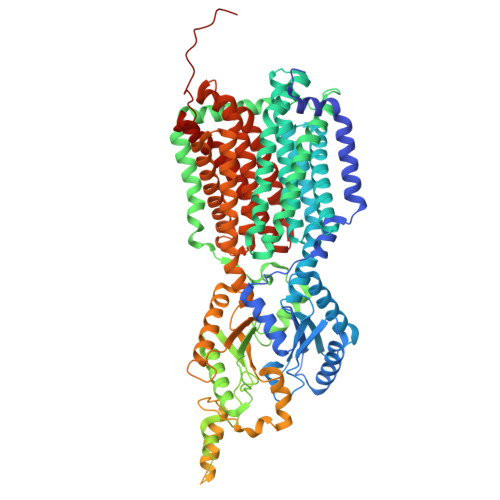 |
UniProt | |||||
Find proteins for P9WJV1 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WJV1 Go to UniProtKB: P9WJV1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WJV1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Siderophore export accessory protein MmpS5 | B [auth F], E [auth C], H [auth G] | 142 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: mmpS5, Rv0677c, MTV040.05c Membrane Entity: Yes | 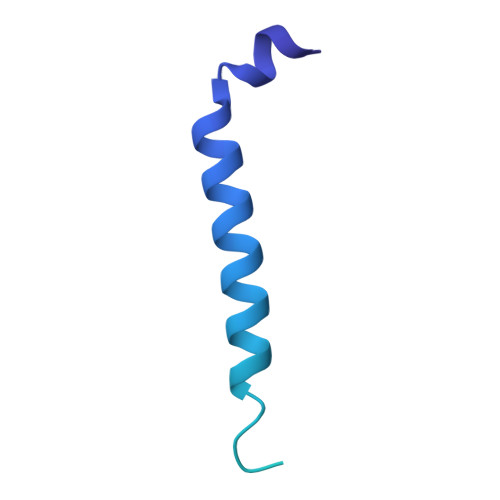 |
UniProt | |||||
Find proteins for P9WJS7 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WJS7 Go to UniProtKB: P9WJS7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WJS7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Meromycolate extension acyl carrier protein | C [auth H], F [auth D], I | 99 | Mycolicibacterium smegmatis MC2 155 | Mutation(s): 0 Gene Names: acpM, MSMEG_4326, MSMEI_4226 | 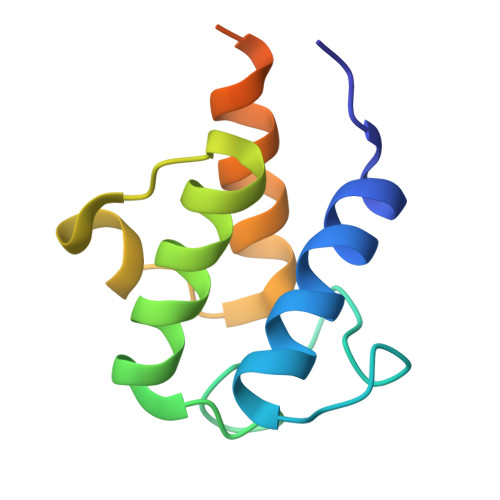 |
UniProt | |||||
Find proteins for A0R0B3 (Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155)) Explore A0R0B3 Go to UniProtKB: A0R0B3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0R0B3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CDL (Subject of Investigation/LOI) Query on CDL | J [auth A] K [auth A] O [auth B] P [auth B] T [auth E] | CARDIOLIPIN C81 H156 O17 P2 XVTUQDWPJJBEHJ-KZCWQMDCSA-L |  | ||
| PEV (Subject of Investigation/LOI) Query on PEV | L [auth A] M [auth A] Q [auth B] R [auth C] V [auth E] | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE C39 H78 N O8 P RPJZYOHZALDGKI-QNGWXLTQSA-N |  | ||
| PN7 (Subject of Investigation/LOI) Query on PN7 | N [auth H], S [auth D], X [auth I] | N~3~-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-(2-sulfanylethyl)-beta-alaninamide C11 H23 N2 O7 P S JDMUPRLRUUMCTL-SECBINFHSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32171217 |
| National Natural Science Foundation of China (NSFC) | China | 32394010 |