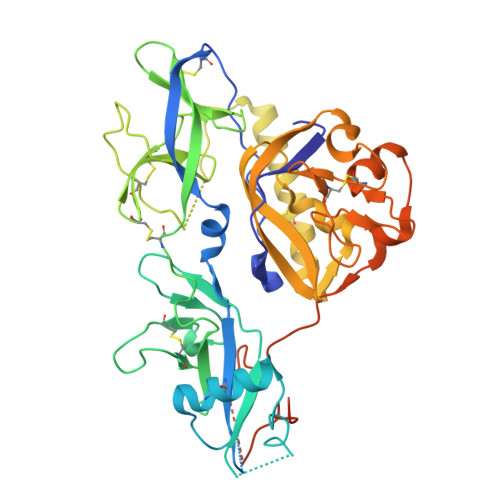
Structures of two lyssavirus glycoproteins trapped in pre- and post-fusion states and the implications on the spatial-temporal conformational transition along with pH-decrease.
Yang, F., Lin, S., Yuan, X., Shu, S., Yu, Y., Yang, J., Ye, F., Chen, Z., He, B., Li, J., Zhao, Q., Ye, H., Cao, Y., Lu, G.(2025) PLoS Pathog 21: e1012923-e1012923
- PubMed: 39970183
- DOI: https://doi.org/10.1371/journal.ppat.1012923
- Primary Citation of Related Structures:
8ZHW, 8ZHZ - PubMed Abstract:
Lyssavirus glycoprotein plays a crucial role in mediating virus entry and serves as the major target for neutralizing antibodies. During membrane fusion, the lyssavirus glycoprotein undergoes a series of low-pH-induced conformational transitions. Here, we report the structures of Ikoma lyssavirus and Mokola lyssavirus glycoproteins, with which we believe that we have trapped the proteins in pre-fusion and post-fusion states respectively. By analyzing the available lyssaviral glycoprotein structures, we present a sequential conformation-transition model, in which two structural elements in the glycoprotein undergo fine-modulated secondary structural transitions, changing the glycoprotein from a bended hairpin conformation to an extended linear conformation. In addition, such conformational change is further facilitated, as observed in our surface plasmon resonance assay, by the pH-regulated interactions between the membrane-proximal region and the pleckstrin homology and the fusion domains. The structural features elucidated in this study will facilitate the design of vaccines and anti-viral drugs against lyssaviruses.
- Department of Emergency Medicine, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, Sichuan, China.
Organizational Affiliation: