Monoclonal humanized monovalent antibody blocking therapy for anti-NMDA receptor encephalitis.
Kanno, A., Kito, T., Maeda, M., Yamaki, S., Amano, Y., Shimomura, T., Anisimova, M., Kanazawa, N., Suzuki, K., Razai, A., Mihara, T., Kubo, K., Shimada, T., Nakamura, K., Nomura, N., Kondo, Y., Okimoto, A., Sugiyama, A., Park, D., Stein, I., Petshow, S., Vandendoren, V., Bilic, S., Kazimi, R., Eastman, V., Snipas, S.J., Mitchell, M., Maurer, M., Jefson, M., Lichter, J., Yamajuku, D., Shirai, H., Adachi, M., Hoeppner, D.J., Kubo, S., Zito, K., Iizuka, T., Flynn, P., Matsumoto, M.(2025) Nat Commun 16: 5292-5292
- PubMed: 40527893
- DOI: https://doi.org/10.1038/s41467-025-60628-1
- Primary Citation of Related Structures:
8ZH7 - PubMed Abstract:
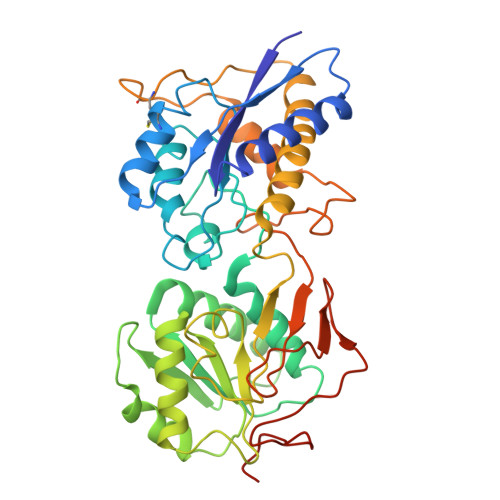
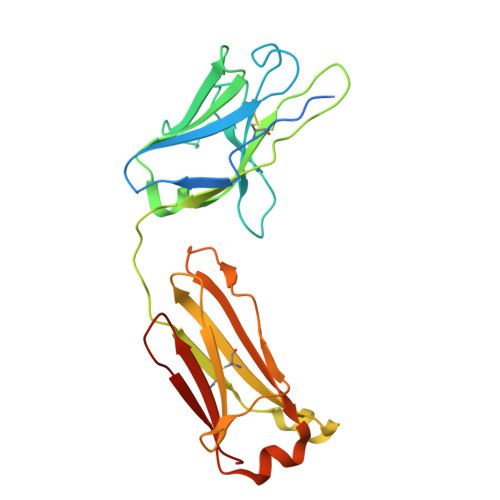
Anti-NMDA receptor (NMDAR) encephalitis is a devastating disease with severe psychiatric and neurological symptoms believed to be caused by pathogenic autoantibodies that bind to the N-terminal domain (NTD) of the NMDAR GluN1 subunit (GluN1-NTD) crosslinking adjacent NMDARs and driving their internalization. Here we describe ART5803, a humanized monovalent antibody, as a potential therapy for anti-NMDAR encephalitis. ART5803 binds with a high affinity (K D = 0.69 nM) to GluN1-NTD without affecting NMDAR activity or inducing internalization. ART5803 blocks NMDAR internalization induced by patients' pathogenic autoantibodies, and restores NMDAR function. A marmoset animal model was developed using sustained intracerebroventricular (ICV) administration of a human pathogenic autoantibody to evoke behavioral and motor abnormalities. ART5803 ICV infusion or peripheral injections rapidly reversed these abnormalities. These data, together with the pharmacokinetic profile in cynomolgus monkeys, indicate a therapeutic potential for intravenous (IV)-administered ART5803 as a fast-acting and efficacious option for anti-NMDAR encephalitis.
- Arialys Therapeutics, Inc., La Jolla, CA, USA.
Organizational Affiliation: