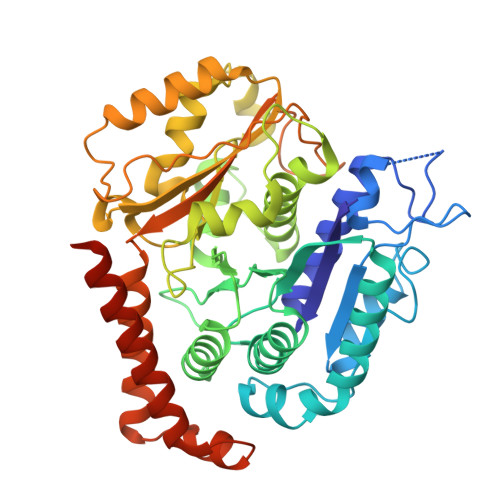
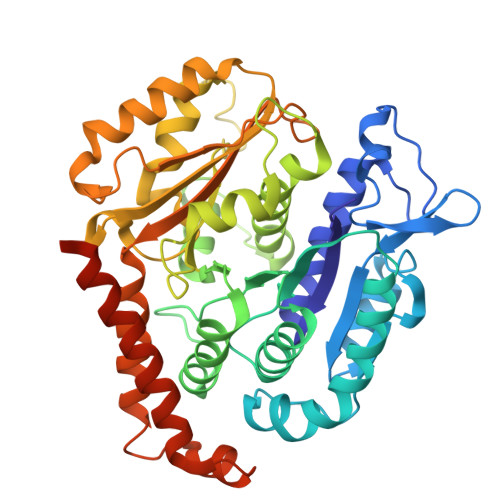
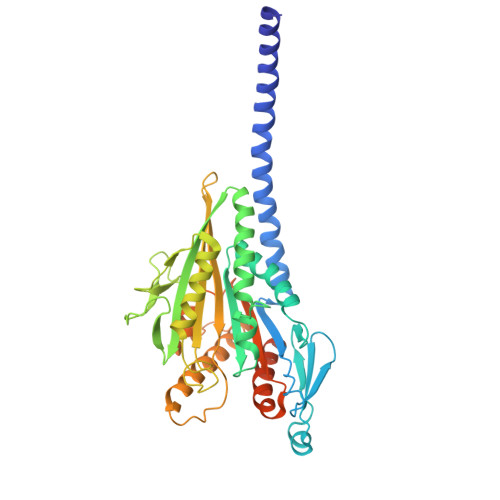
Structural transitions in kinesin minus-end directed microtubule motility.
Shibata, S., Wang, M.Y., Imasaki, T., Shigematsu, H., Wei, Y., Jobichen, C., Hagio, H., Sivaraman, J., Endow, S.A., Nitta, R.(2024) bioRxiv
- PubMed: 39131399
- DOI: https://doi.org/10.1101/2024.07.29.605428
- Primary Citation of Related Structures:
8YY2, 8YY3, 8YY4, 8YY5 - PubMed Abstract:
Kinesin motor proteins hydrolyze ATP to produce force for spindle assembly and vesicle transport, performing essential functions in cell division and motility, but the structural changes required for force generation are uncertain. We now report high-resolution structures showing new transitions in the kinesin mechanochemical cycle, including power stroke fluctuations upon ATP binding and a post-hydrolysis state with bound ADP + free phosphate. We find that rate-limiting ADP release occurs upon microtubule binding, accompanied by central β-sheet twisting, which triggers the power stroke - stalk rotation and neck mimic docking - upon ATP binding. Microtubule release occurs with β-strand-to-loop transitions, implying that β-strand refolding induces Pi release and the recovery stroke. The strained β-sheet during the power stroke and strand-to-loop transitions identify the β-sheet as the long-sought motor spring.
- Division of Structural Medicine and Anatomy, Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, 650-0017, Japan.
Organizational Affiliation: