Molecular mechanism and structure-guided humanization of a broadly neutralizing antibody against SFTSV.
Yang, P., Wu, X., Shang, H., Sun, Z., Wang, Z., Song, Z., Yuan, H., Deng, F., Shen, S., Guo, Y., Zhang, N.(2024) PLoS Pathog 20: e1012550-e1012550
- PubMed: 39321193
- DOI: https://doi.org/10.1371/journal.ppat.1012550
- Primary Citation of Related Structures:
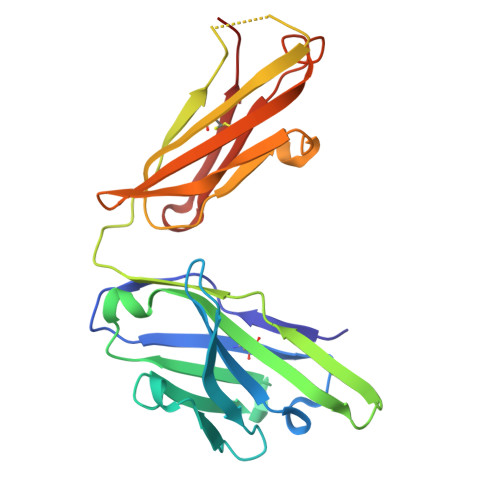
8YXI - PubMed Abstract:
Severe fever with thrombocytopenia syndrome virus (SFTSV) is a novel tick-borne bunyavirus that causes severe fever with thrombocytopenia syndrome (SFTS), with a high mortality rate of up to 30%. The envelope glycoproteins of SFTSV, glycoprotein N (Gn) and glycoprotein C (Gc), facilitate the recognition of host receptors and the process of membrane fusion, allowing the virus to enter host cells. We previously reported a monoclonal antibody, mAb 40C10, capable of neutralizing different genotypes of SFTSV and SFTSV-related viruses. However, the specific neutralization mechanism is poorly understood. In this study, we elucidated the high-resolution structure of the SFTSV Gn head domain in complex with mAb 40C10, confirming that the binding epitope in the domain I region of SFTSV Gn, and it represented that a novel binding epitope of SFTSV Gn was identified. Through in-depth structural and sequence analyses, we found that the binding sites of mAb 40C10 are relatively conserved among different genotypes of SFTSV and SFTSV-related Heartland virus and Guertu virus, elucidating the molecular mechanism underlying the broad-spectrum neutralizing activity of mAb 40C10. Furthermore, we humanized of mAb 40C10, which is originally of murine origin, to reduce its immunogenicity. The resulting nine humanized antibodies maintained potent affinity and neutralizing activity. One of the humanized antibodies exhibited neutralizing activity at picomolar IC50 values and demonstrated effective therapeutic and protective effects in a mouse infection model. These findings provide a novel target for the future development of SFTSV vaccines or drugs and establish a foundation for the research and development of antibody therapeutics for clinical applications.
- State Key Laboratory of Medicinal Chemical Biology and College of Life Sciences, Nankai University, Tianjin, China.
Organizational Affiliation: